
An introduction to image processing using ImageJ
Mark Willett, Imaging and Microscopy Centre,
Centre for Biological Sciences, University of Southampton.
1

[Shift] + Left click to select and open
everything between two selections in a list
[Ctrl] + Left click to make multiple
individual selections
Drag and drop onto the menu bar to open single or multiple images or stacks
2

A note on image formats
Images are comprised of pixels, each with an x,y coordinate and an intensity value that gives
the pixel contrast in comparison to adjacent pixels.
The bit-depth of an image is defined by the number of intensity values available to each of
the pixels in an image.
e.g.
Binary = 2 values, (black or white)
8-bit = 256 values. Black (0), white (255) and a scale of 254 grey steps in between
12-bit = 4,096 grey values
16-bit = 65,536 grey values
RGB formats are colour images comprised of 3 channels (red, green and blue) each channel
has independent grey values, usually 8-bit
e.g. an RGB image that has 3 channels x 256 possible values per pixel is 3 x 8-bit = 24-bit
3

The standard scientific image file format is the uncompressed Tagged Image File Format (.tiff,
.tif) although some microscope manufacturers use their own similar proprietary lossless
formats.
.Tiffs contain an embedded notepad file that contains essential information about the image
such as scaling, laser wavelengths etc (metadata).
Non-scientific file formats such as .jpeg should be avoided as they are “lossy” (some of the
image information is lost because they are compressed to reduce the file size, but are usually
ok for presentations).
It’s usually also best to avoid Windows art packages and non-scientific image viewing and
manipulation packages as they may apply compression to your image and display your image
differently from scientific viewing and analysis packages.
A note on image formats
4

5
Viewing image metadata (.tiffs only)
View image metadata by selecting Image>Show Info... Metadata contains
information such as scaling, pixel dimensions, bit depth, fluorescence wavelength
etc.

Some image analysis functions need to use 8-bit image formats (256 grey values)
Change the bit depth using Image>Type.
6

7
0
255
Dynamic range of grey values
Number of pixels
Displayed
black value
Displayed
white value
Distribution of
image intensity
values
The image histogram (8-bit image example)

8
0
255
Number of pixels
Maximum pixel value is 80 out of a possible 255. The image is too
dark. This might have been necessary to retain a short exposure
time or reduce photobleaching/phototoxicity of the specimen.

9
0
255
Number of pixels
80
Image is bright but only has
80 steps of intensity
Intensity scaling
When the white value is moved all of the grey values between black and white are re-
scaled so the image appears brighter, however actual pixel values remain unchanged.

10
Number of pixels
All pixels set to
white
255
Image has been saturated during acquisition. Contrast of all
pixels over 255 is permanently lost

11
0
255
Number of pixels
All pixels set to
white
200
Image saturated during processing by incorrect placement of white value

12
0
255
Number of pixels
All pixels
set to black
45
Removes background (and low intensity image information) noise or tell ImageJ
which intensities to send to black and which to white when making a binary image.
Thresholding

13
Scaling image brightness automatically
Open image “Microtubules 8-bit”. This image does not use the whole dynamic range.
Image>Adjust>Brightness/Contrast, Select ”Auto”. Don’t adjust sliders.
This function moves the displayed white value to the point where 0.4% of pixels are
saturated. All the grey values are then re-scaled and the image appears brighter .
As you can see from the histogram, actual pixel values remain unchanged and intensity
can still be measured if desired.
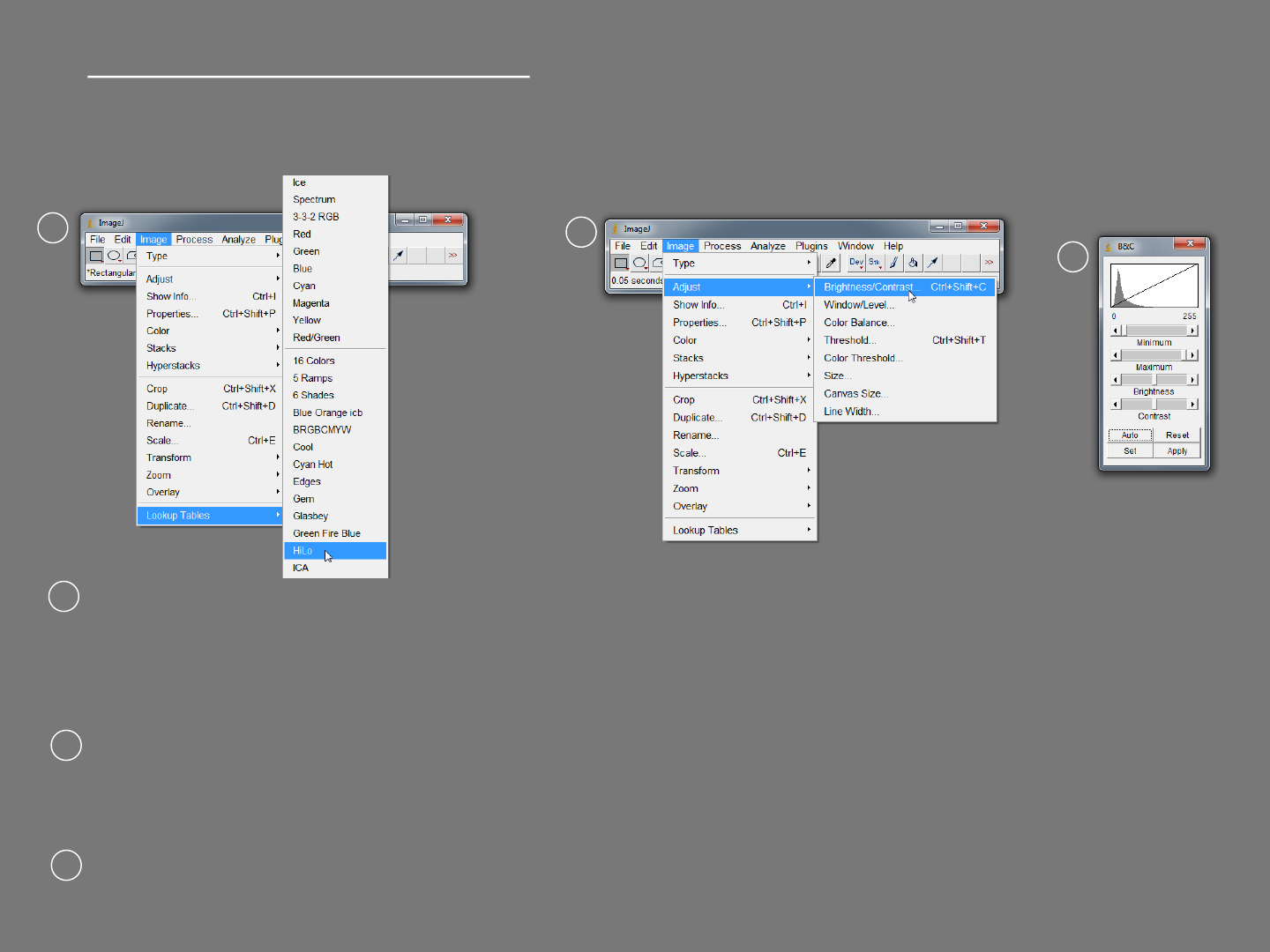
Scaling image brightness manually
Open “Microtubules 8-bit” again. This method also does not change actual pixel
values.
Select Image>Lookup Tables>HiLo and then Image>Adjust>Brightness/Contrast.
Saturated pixels (value of 255) now appear red and pixels with a value of 0 appear
blue.
Adjust the Maximum slider until you get few red pixels and then back it off until they
just disappear.
Adjust the “Minimum” slider until the background turns blue. Click “Apply” then
select Image>Lookup Tables>Grays.
1
2
3
1
2
3
14

15
1
2
3
1
3
2
Open image “Peccary hair”
Select Image>Transform>Rotate....
Tick “Preview”. Adjust Angle slider until to
achieve the desired rotation. Click OK.
Rotating images

16
1
2
Cropping
3
1
2
With the image you just rotated, using
the rectangular selection icon, drag a
selection around the area that you
want to keep.
Select Image>Crop
For consistency of size, crop regions can be
stored in the ROI mananger.
Analyze>Tools>ROI Manager...

Calibrating images
Before you can add a scale bar or analyse images, the images have to be calibrated to the
correct measurement units.
Many instruments automatically add spatial calibrations to the image metadata during
acquisition. To check if your images are calibrated look in the top left hand corner of the
image. If your image dimensions are given in pixel units the image has not been
calibrated.
Uncalibrated
Calibrated
If your image is not automatically calibrated by the acquisition software, an image of a
stage micrometer taken at the same magnification as your specimen can be used to
calibrate your images.
17

(1 small increment on the micrometer = 10 µm)
1
2
3
Open images “calibration image” and
“Uncalibrated image”. Using the line tool,
draw a line of a known distance on the
image of the stage micrometer.
Select Analyze>Set Scale.
Enter the known distance and units in
um. Ticking “Global” applies the
calibration to all images in the Imagej
session.
Manually calibrating images
18

Adding a scale bar
With your calibrated image open
select Analyze>Tools>Scale Bar.
Select the required width and
position and click “OK”.
19

Merging images into multichannel 24-bit RGB .tifs
Open images “Blue.tif, Green.tif, Red.tif” and “Brightfield.tif”.
Select Image>Colour>Merge Channels
Select each image into their corresponding colour channels.
Untick “Create composite”, tick “Keep source images” and click “OK”.
Select Image>Colour>Merge Channels
Select Green.tif in the green channel and Brightfield.tif in the grey channel
Untick “Create composite”, tick “Keep source images” and click “OK”.
1
2
1
2
20

Splitting multichannel RGB images
Open image “Mitotic cell”.
Select Image>Colour>Split Channels.
21

Applying pseudocolour to grey images
Using your open grey images split from “Mitotic cell.tif”, Select a channel image by
clicking on it then select Image>Lookup Tables.
Select the correct corresponding colour from the drop down menu.
To return a channel to grey, select “Grays” from the drop down menu.
22

Stacks
y
x
z or t
Individual
images
Stacks are a method of handling multiple related images in one file.
They are often used to handle multiple slices through the vertical z-axis of a specimen
(a z-stack), but are also used to handle sequential images in a time lapse experiment (a t-
stack) or images acquired at different wavelengths (a λ-stack).
Stacks can have up to three dimensions e.g. [x,y],[channels],[z or t or λ].
Stacks with more than three dimensions e.g. {[x,y],[channels],[z]},[t] are called hyperstacks.
23

View image stacks as a montage
Open z-stack 4EBP1.tif
Select Image>Stacks>Make Montage
24

Convert stacks to single images
Open z-stack 4EBP1.tif
Select Image>Stacks>Stack to Images
25

Convert single images to stacks
Using your single images split from z-stack 4EBP1.tif
Select Image>Stacks>Images to Stack
In the pop up window, type something that is in the title of all of the images.
26

27
Make a substack or a single slice from a stack into a separate image
Displayed Slice number is shown in the top left hand corner of the stack
Select Image>Stacks>Tools>Make Substack
Enter slice number for a single image, or range if you want to make a substack.

28
Project a stack to a 2D image
Select Image>Stacks>Z Project….
Select start and finish slices that you want to project and Projection type “Max intensity”

Select regions of interest (ROIs) for measurement using the Rectangular, Polygon, Oval,
Freehand and Line selection tools.
When the cursor is a cross you can click and drag to make your ROI. Placing your cursor in the
middle of your ROI (it changes to an arrow) allows you to move your ROI.
Open image 4E Cisternae.tif
Basic selections and measurements
29

Basic selections and measurements
To set the type of measurement to be made with the ROI, select
Analyse>Set Measurements... and tick the relevant boxes then click “OK”.
Area returns the area within the ROI in the calibrated units
2
(or pixels
2
if the image is
uncalibrated).
Mean gray value returns the average grey value inside the ROI.
Integrated density is the average grey value * area inside the ROI.
30
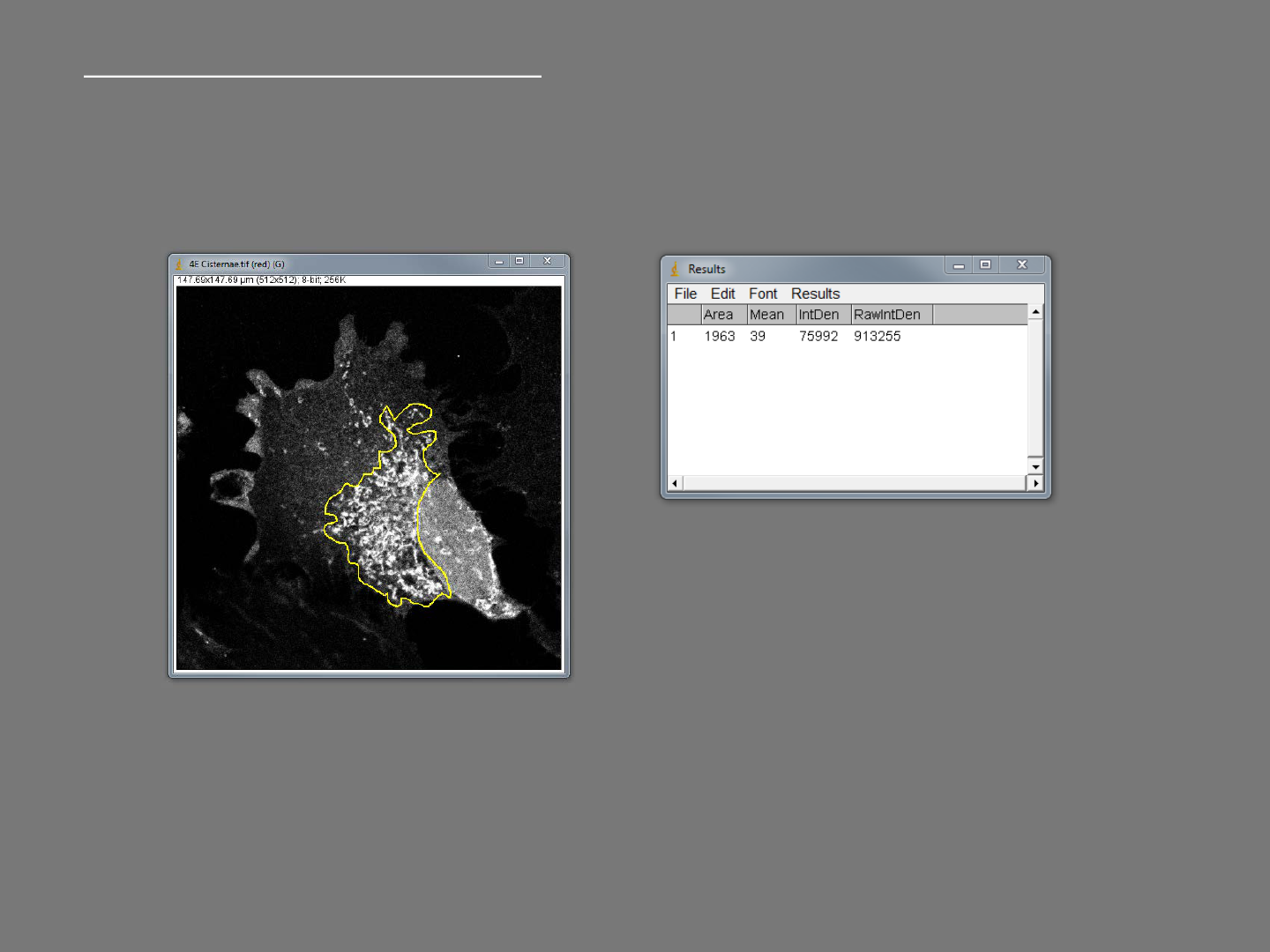
Basic selections and measurements
Press [Ctrl+M] to make the measurement
Draw an ROI on the image using one of the selection tools.
31

Multiple measurements can be made (select a
new ROI and press [Ctrl+M] again).
You can calculate the mean and standard
deviation of the whole dataset by selecting
Results>Summarize from the Results window.
Results can also be copied and pasted into
Excel for further analysis.
Basic selections and measurements
32

The ROI manager
Multiple ROIs can be stored and recalled using
the ROI manager.
Select Analyze>Tools>ROI Manager...
To add the current ROI to the ROI manager
click “Add”.
To recall the ROI into any image select it from
the list on the left.
Select More>>Save... On the ROI manager
window to save ROIs for later use.
To change the colour and weight of the ROI
click “Properties” on the ROI manager window.
33

Background correction
Ideally the grey value for the image background should be as close to 0 as possible
with similar values across the whole image background.
Poor quality or incorrectly configured illumination may cause an uneven background.
Some microscope systems allow grey value “offset” during acquisition, however this
can also be achieved post-acquisition if necessary using several methods:
Rolling Ball – Corrects uneven illumination and preserves low intensity fine specimen
detail.
Thresholding – Subjective and removes low intensity image information as well as
the background.
Background subtraction – Removes low intensity image information as well as the
background. Based on average background intensity, better when quantification is
required.
34

Background correction - Rolling Ball
This method Imagines a ball or paraboloid rolled around the image. Areas of grey
value large enough for the “ball” to touch to are removed.
This has the advantage over simple background subtraction of preserving low
intensity fine detail in the image and correcting uneven illumination.
Before After
35
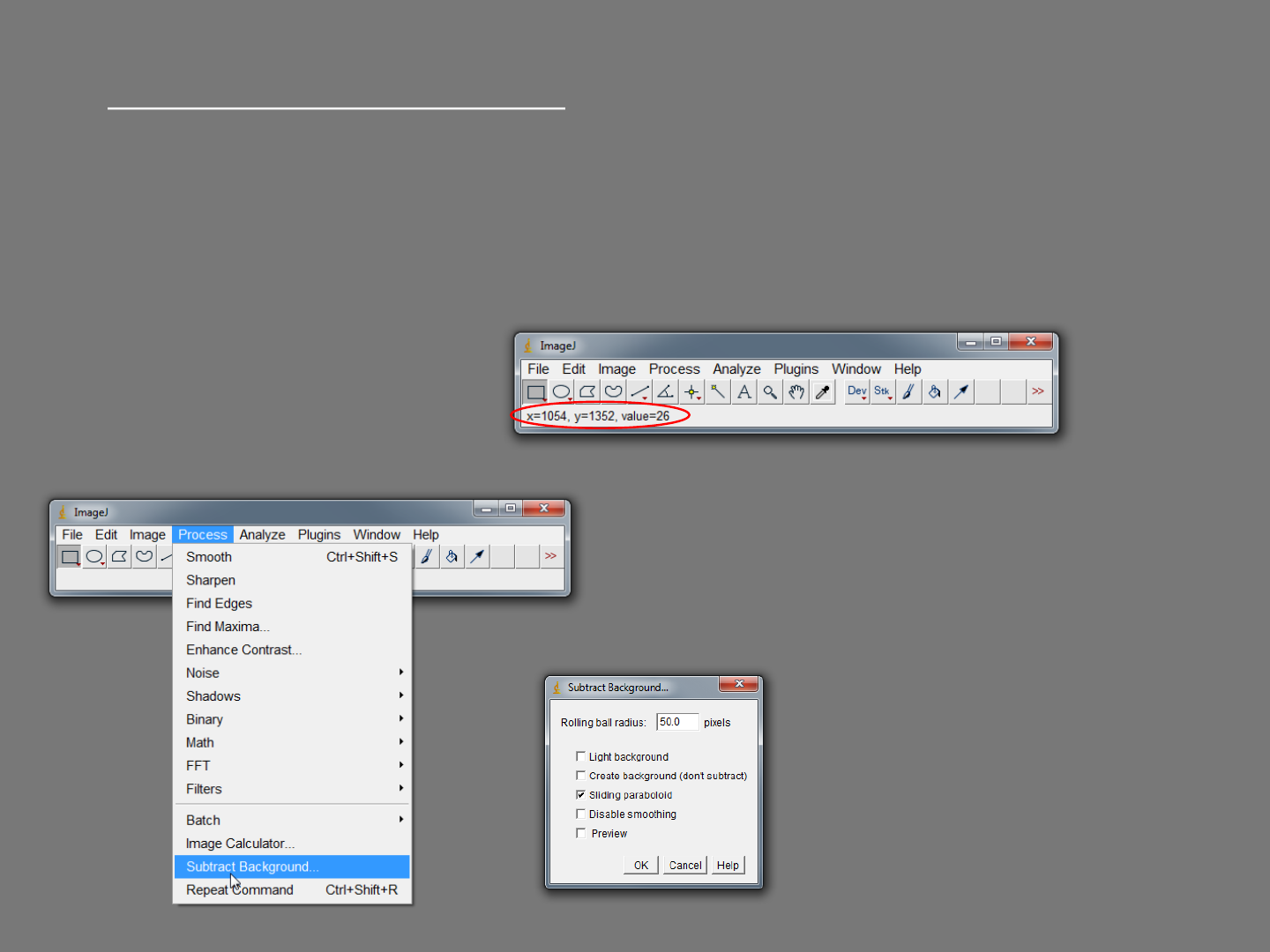
Background correction - Rolling Ball
Open image “Background correction example”. This image has a background
problem as well as uneven illumination, and has a large circular artefact from the
dish that the cells were imaged in.
If you hover the cursor over the image, the grey value of the pixel under the cursor is
displayed on the menu bar.
Select Process>Subtract Background. Tick
“Sliding paraboloid” and choose a “Rolling
ball radius” of 50 pixels and click “OK”.
36

Background correction - Thresholding
Close your background corrected image without saving and open image “Background
correction example” again.
Select Image>Lookup Tables>HiLo and then Image>Adjust>Brightness/Contrast.
Saturated pixels (value of 255) now appear red and pixels with a value of 0 appear blue.
Adjust the “Minimum” slider until the entire background is blue. All pixels with a value
below the selected threshold (including image information!) will be set to 0. Click “Apply”
then Select Image>Lookup Tables>Grays.
1 2
3
37

Background correction - Subtraction
Close your background corrected image without saving and open image “Background
correction example” again.
Select Analyze>Set Measurements and then tick the “Mean gray value” box and click
“OK”.
Select an ROI on an area of background and select Analyze>Measure or use [Ctrl+M]. A
results window should appear with the mean grey value of the pixels within your ROI.
38

Background correction - Subtraction
Deselect your ROI by clicking on the image or the subtraction will only be applied within
the ROI. Select Process>Math>Subtract and enter your mean background value into the
value field on the window that appears. Click “OK“ or select “Preview”.
39

Removing noise
Image noise tends to manifest as speckle and can be caused by:
Electronic variations in imaging detectors.
Analogue to digital conversion during image acquisition.
Variations in photon detection, particularly from low signal specimens: “shot noise”.
High detector gain: “dark noise”.
Speckle can also be caused by contamination such as dust in the sample auto-fluorescing
and by precipitates from stains, so its good practice to wash or flame coverslips and slides
and centrifuge stains prior to use.
Care should be taken when doing noise corrections on images, which may not be suitable
if quantitative measurements are to be made.
40

Removing noise - outliers
Open image “despeckle 3”. This image is suffering from speckle caused by high detector
gain and also has some large speckles which are probably caused by stain precipitates
reacting with the coverslip coating.
Remove the large bright speckles:
This function replaces a pixel with the median value of the surrounding pixel values if it
deviates from the median by more than the thresholded value. Select “Preview” Reduce
the threshold value until the bright outliers disappear (about 90 in this case). Click “OK”
when finished
41

Removing noise - speckle
Remove the detector noise:
This function replaces each pixel with the median value of the surrounding 3x3 pixels.
Select Process>Noise>Despeckle.
Use image>Adjust>Brightness/Contrast and the minimum slider to background correct
your image
Open the original image to compare the differences.
42

Making precise selections – binary selection method
Open image “C elegans lipid store”.
Set the second pulldown menu to “Red”.
Select Image>Adjust>Threshold and threshold the image using the “minimum” slider.
Click “Apply” to make a binary image.
43

Making precise selections – binary selection method
Select Edit>Selection>Create Selection. An ROI will appear over the binary image
44

Making precise selections – binary selection method
Select Analyze>Tools>ROI Manager and click “Add”. A new ROI appears in the ROI
manager.
Open the original image “C elegans lipid store”. Select the ROI in the ROI manager, the
ROI will appear on the original image ready for analysis.
1
2
1
2
45

Making precise selections – Wand (tracing) tool
The wand (tracing) tool Makes selections
based on grey values when clicked or
dragged across an image.
You can double click the icon to set the tolerance, but it works best
with a thresholded or binary image.
Open “Mitotic cell.tif (red)”.
Select Image>Adjust>Threshold and threshold the image.
Do not click “Apply”.
46

Select the Wand (tracing) tool. Click
and drag to make the selection
then add your selection to the ROI
manager.
or reset the threshold
And make your
measurements immediately
Making precise selections – Wand (tracing) tool
47
