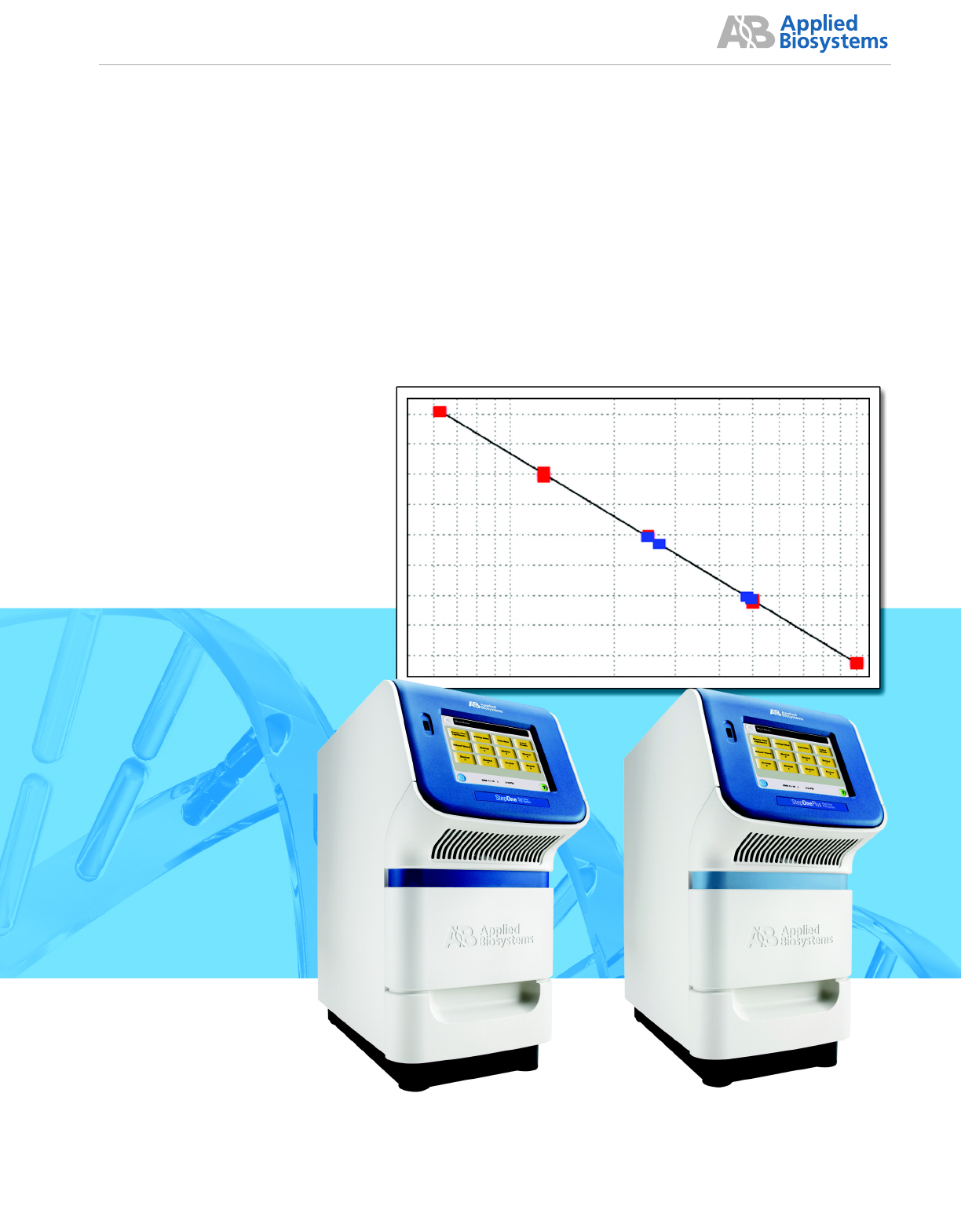
Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems
Standard Curve Experiments
Getting Started Guide
© Copyright 2008, 2010 Applied Biosystems. All rights reserved.
Information in this document is subject to change without notice. Applied Biosystems assumes no responsibility for any errors that may appear in this
document.
APPLIED BIOSYSTEMS DISCLAIMS ALL WARRANTIES WITH RESPECT TO THIS DOCUMENT, EXPRESSED OR IMPLIED, INCLUDING BUT
NOT LIMITED TO THOSE OF MERCHANTABILITY OR FITNESS FOR A PARTICULAR PURPOSE. IN NO EVENT SHALL APPLIED
BIOSYSTEMS BE LIABLE, WHETHER IN CONTRACT, TORT, WARRANTY, OR UNDER ANY STATUTE OR ON ANY OTHER BASIS FOR
SPECIAL, INCIDENTAL, INDIRECT, PUNITIVE, MULTIPLE OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR ARISING FROM
THIS DOCUMENT, INCLUDING BUT NOT LIMITED TO THE USE THEREOF.
For Research U se Only. Not for use in diagnostic procedures.
NOTICE TO PURCHASER: Label License
The StepOne
™
and StepOnePlus
™
Real-Time PCR Systems are covered by US patents and corresponding claims in their non-US counterparts, owned by
Applied Biosystems Inc. No right is conveyed expressly, by implication, or by estoppel under any other patent claim, such as claims to apparatus,
reagents, kits, or methods such as 5′ nuclease methods. Further information on purchasing licenses may be obtained by contacting the Director of
Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.
TRADEMARKS:
Applied Biosystems, AB (Design), MicroAmp, Primer Express, and VIC are registered trademarks, and FAM, JOE, ROX, StepOne, StepOnePlus, TAMRA,
and VeriFlex are trademarks of Applied Biosystems or its subsidiaries in the U.S. and/or certain other countries.
AmpErase, AmpliTaq Gold, and TaqMan are registered trademarks of Roche Molecular Systems, Inc.
SYBR is a registered trademark of Molecular Probes, Inc.
Macintosh is a registered trademark of Apple Computer, Inc.
Microsoft and Windows are registered trademarks of Microsoft Corporation.
All other trademarks are the sole property of their respective owners.
Part Number 4376784 Rev. F
08/2010
Contents
iii
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Preface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . vii
How to Use This Guide . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . vii
How to Obtain More Information . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ix
How to Obtain Support . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xii
Safety Conventions Used in This Document . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xiii
Symbols on Instruments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xiv
Safety Labels on Instruments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xvi
General Instrument Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xvii
Chemical Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xviii
Chemical Waste Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xix
Electrical Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xx
LED Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xxi
Biological Hazard Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xxi
Workstation Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . xxii
Safety and Electromagnetic Compatibility (EMC) Standards . . . . . . . . . . . . . . . . . . . xxii
Chapter 1 Get Started. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1
About the StepOne
™
and StepOnePlus
™
Systems . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2
Supported Consumables . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 4
About Standard Curve Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
How to Use This Guide . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
About the Example Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
Example Experiment Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12
Chapter 2 Design the Experiment . . . . . . . . . . . . . . . . . . . . . . . . 15
Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16
Create a New Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
Define the Experiment Properties . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20
Define the Methods and Materials . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22
Set Up the Targets . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25
Set Up the Standards . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27
Set Up the Samples . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29
Set Up the Run Method . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 31
Review the Reaction Setup . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33
Order Materials for the Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 38
Finish the Design Wizard . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 42
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
iv
Chapter 3 Prepare the Reactions . . . . . . . . . . . . . . . . . . . . . . . . . 45
Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 46
Prepare the Sample Dilutions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 47
Prepare the Standard Dilution Series . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 49
Prepare the Reaction Mix . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 51
Prepare the Reaction Plate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 53
Chapter 4 Run the Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . 59
Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 60
Prepare for the Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 61
(Optional) Enable the Notification Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 63
Start the Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 65
Monitor the Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 69
Unload the Instrument and Transfer the Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 76
Chapter 5 Analyze the Experiment . . . . . . . . . . . . . . . . . . . . . . . . 81
Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 82
Section 5.1: Review Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83
Analyze the Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84
View the Standard Curve . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 90
View the Amplification Plot . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 92
View the Well Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 99
Publish the Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 102
Section 5.2: Troubleshoot (If Needed) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 103
View the Analysis Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 104
View the QC Summary . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 106
Omit Wells from the Analysis . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 108
View the Multicomponent Plot . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 109
View the Raw Data Plot . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 112
Appendix A Alternate Experiment Workflows . . . . . . . . . . . . . . . . . . 115
Advanced Setup Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 116
QuickStart Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 117
Template Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 119
Export/Import Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 121
v
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Bibliography . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 125
Glossary . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 127
Index . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 143
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
vi

vii
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Preface
How to Use This Guide
About the System
Documentation
The guides listed below are shipped with the Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems (StepOne
™
and StepOnePlus
™
systems).
Guide Purpose and Audience PN
Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems
Getting Started Guide for Genotyping
Experiments
Explains how to perform experiments on the StepOne and
StepOnePlus systems. Each Getting Started Guide functions as
both:
• A tutorial, using example experiment data provided with the
Applied Biosystems StepOne
™
Real-Time PCR Software
(StepOne
™
software).
• A guide for your own experiments.
Intended for laboratory staff and principal investigators who
perform experiments using the StepOne or StepOnePlus
system.
4376786
Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems
Getting Started Guide for
Presence/Absence Experiments
4376787
Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems
Getting Started Guide for Relative
Standard Curve and Comparative C
T
Experiments
4376785
Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems
Getting Started Guide for Standard Curve
Experiments
4376784
Applied Biosystems StepOne
™
and
StepOnePlus™ Real-Time PCR Systems
Installation, Networking, and Maintenance
Guide
Explains how to install and maintain the StepOne and
StepOnePlus systems.
Intended for laboratory staff responsible for the installation and
maintenance of the StepOne or StepOnePlus system
4376782
Applied Biosystems StepOne
™
and
StepOnePlus™ Real-Time PCR Systems
Installation Quick Reference Card
4376783
Applied Biosystems StepOne
™
and
StepOnePlus™ Real-Time PCR Systems
Reagent Guide
Provides information about the reagents you can use on the
StepOne and StepOnePlus systems, including:
• An introduction to TaqMan
®
and SYBR
®
Green reagents
• Descriptions and design guidelines for the following
experiment types:
– Quantitation experiments
– Genotyping experiments
– Presence/absence experiments
Intended for laboratory staff and principal investigators who
perform experiments using the StepOne or StepOnePlus
system.
4379704

Preface
How to Use This Guide
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
viii
Assumptions
This guide assumes that you:
• Are familiar with the Microsoft Windows
®
XP operating system.
• Are familiar with the Internet and Internet browsers.
• Know how to handle DNA and/or RNA samples and prepare them for PCR.
• Understand data storage, file transfer, and copying and pasting.
• Have networking experience, if you plan to integrate the StepOne or StepOnePlus
system into your existing laboratory data flow.
Te xt Co n v e ntio n s
This guide uses the following conventions:
• Bold text indicates user action. For example:
Ty pe 0, then press Enter for each of the remaining fields.
• Italic text indicates new or important words and is also used for emphasis.
For example:
Before analyzing, always prepare fresh matrix.
• A right arrow symbol () separates successive commands you select from a drop-
down or shortcut menu. For example:
Select FileOpen.
User Attention
Words
Two user attention words appear in Applied Biosystems user documentation. Each word
implies a particular level of observation or action as described below:
Note: – Provides information that may be of interest or help but is not critical to the use
of the product.
IMPORTANT! – Provides information that is necessary for proper instrument operation,
accurate reagent kit use, or safe use of a chemical.
Applied Biosystems StepOne
™
and
StepOnePlus™ Real-Time PCR Systems
Site Preparation Guide
Explains how to prepare your site to receive and install the
StepOne and StepOnePlus systems.
Intended for personnel who schedule, manage, and perform the
tasks required to prepare your site for installation of the StepOne
or StepOnePlus system.
4376768
Applied Biosystems StepOne
™
Real-Time
PCR Software Help
Explains how to use the StepOne software to:
• Set up, run, and analyze experiments using the StepOne and
StepOnePlus systems.
• Monitor networked StepOne and StepOnePlus instruments.
• Calibrate StepOne and StepOnePlus instruments.
• Verify the performance of StepOne and StepOnePlus
instruments with an RNase P run.
Intended for:
• Laboratory staff and principal investigators who perform
experiments using the StepOne or StepOnePlus system.
• Laboratory staff responsible for the installation and
maintenance of the StepOne or StepOnePlus system.
NA
Guide Purpose and Audience PN

Preface
How to Obtain More Information
ix
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Examples of the user attention words appear below:
Note: The Calibrate function is also available in the Control Console.
IMPORTANT! To verify your client connection, you need a valid user ID.
Safety Alert
Words
Safety alert words also appear in user documentation. For more information, see “Safety
Alert Words” on page xiii.
How to Obtain More Information
Related
Documentation
Other StepOne and StepOnePlus System Documents
The documents listed in the table below are not shipped with the StepOne or
StepOnePlus instrument.
Documents Related to Genotyping Experiments
Document PN
Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems
Installation Performance Verification Protocol
4376791
Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems
Installation Qualification-Operation Qualification Protocol
4376790
Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems
Planned Maintenance Protocol
4376788
Document PN
Allelic Discrimination Pre-Developed TaqMan
®
Assay Reagents Quick Reference
Card
4312212
Custom TaqMan
®
Genomic Assays Protocol 4367671
Custom TaqMan
®
SNP Genotyping Assays Protocol 4334431
Ordering TaqMan
®
SNP Genotyping Assays Quick Reference Card 4374204
Performing a Custom TaqMan
®
SNP Genotyping Assay for 96-Well Plates Quick
Reference Card
4371394
Performing a TaqMan
®
Drug Metabolism Genotyping Assay for 96-Well Plates
Quick Reference Card
4367636
Pre-Developed TaqMan
®
Assay Reagents Allelic Discrimination Protocol 4312214
TaqMan
®
Drug Metabolism Genotyping Assays Protocol 4362038
TaqMan
®
SNP Genotyping Assays Protocol 4332856

Preface
How to Obtain More Information
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
x
Documents Related to Presence/Absence Experiments
Documents Related to Relative Standard Curve and Comparative C
T
Experiments
Documents Related to Standard Curve Experiments
Document PN
DNA Isolation from Fresh and Frozen Blood, Tissue Culture Cells, and Buccal
Swabs Protocol
4343586
NucPrep
®
Chemistry: Isolation of Genomic DNA from Animal and Plant Tissue
Protocol
4333959
PrepMan
®
Ultra Sample Preparation Reagent Protocol 4318925
Document PN
Amplification Efficiency of TaqMan
®
Gene Expression Assays Application Note 127AP05
Applied Biosystems High-Capacity cDNA Reverse Transcription Kits Protocol 4375575
Custom TaqMan
®
Gene Expression Assays Protocol 4334429
Primer Express
®
Software Version 3.0 Getting Started Guide 4362460
TaqMan
®
Gene Expression Assays Protocol 4333458
User Bulletin #2: Relative Quantitation of Gene Expression 4303859
Document PN
Amplification Efficiency of TaqMan
®
Gene Expression Assays Application Note 127AP05
Custom TaqMan
®
Gene Expression Assays Protocol 4334429
Primer Express
®
Software Version 3.0 Getting Started Guide 4362460
TaqMan
®
Gene Expression Assays Protocol 4333458
User Bulletin #2: Relative Quantitation of Gene Expression 4303859

Preface
How to Obtain More Information
xi
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Documents Related to the Reagent Guide
Note: For more documentation, see “How to Obtain Support” on page xii.
Obtaining
Information from
the Software Help
The StepOne Software Help describes how to use each feature of the user interface.
Access the Help from within the software by doing one of the following:
•Press F1.
• Click in the toolbar.
• Select HelpStepOne Software Help.
To find topics of interest in the Help:
• Review the table of contents.
• Search for a specific topic.
• Search an alphabetized index.
Document PN
Applied Biosystems High-Capacity cDNA Reverse Transcription Kits Protocol 4375575
Custom TaqMan
®
Gene Expression Assays Protocol 4334429
Custom TaqMan
®
Genomic Assays Protocol: Submission Guidelines 4367671
Custom TaqMan
®
SNP Genotyping Assays Protocol 4334431
Power SYBR
®
Green PCR Master Mix and RT-PCR Protocol 4367218
Pre-Developed TaqMan
®
Assay Reagents Allelic Discrimination Protocol 4312214
Primer Express
®
Software Version 3.0 Getting Started Guide 4362460
SYBR
®
Green PCR and RT-PCR Reagents Protocol 4304965
SYBR
®
Green PCR Master Mix and RT-PCR Reagents Protocol 4310251
TaqMan
®
Drug Metabolism Genotyping Assays Protocol 4362038
TaqMan
®
Exogenous Internal Positive Control Reagents Protocol 4308335
TaqMan
®
Fast Universal PCR Master Mix (2✕) Protocol 4351891
TaqMan
®
Gene Expression Assays Protocol 4333458
TaqMan
®
Gene Expression Master Mix Protocol 4371135
TaqMan
®
Genotyping Master Mix Protocol 4371131
TaqMan
®
SNP Genotyping Assays Protocol 4332856
TaqMan
®
Universal PCR Master Mix Protocol 4304449
User Bulletin #2: Relative Quantitation of Gene Expression 4303859
Using TaqMan
®
Endogenous Control Assays to Select an Endogenous Control
for Experimental Studies Application Note
127AP08

Preface
How to Obtain Support
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
xii
Send Us Your
Comments
Applied Biosystems welcomes your comments and suggestions for improving its user
documents. You can e-mail your comments to:
techpubs@appliedbiosystems.com
IMPORTANT! The e-mail address above is only for submitting comments and
suggestions relating to documentation. To order documents, download PDF files, or for
help with a technical question, go to http://www.appliedbiosystems.com, then click the
link for Support. (See “How to Obtain Support” on page xii).
How to Obtain Support
For the latest services and support information for all locations, go to
http://www.appliedbiosystems.com, then click the link for Support.
At the Support page, you can:
• Search through frequently asked questions (FAQs)
• Submit a question directly to Technical Support
• Order Applied Biosystems user documents, MSDSs, certificates of analysis, and
other related documents
• Download PDF documents
• Obtain information about customer training
• Download software updates and patches
In addition, the Support page provides access to worldwide telephone and fax numbers
to contact Applied Biosystems Technical Support and Sales facilities.
IMPORTANT! When directed to do so by this guide, or when you need to schedule
maintenance for your StepOne
™
or StepOnePlus
™
instrument (such as annual planned
maintenance or temperature verification/calibration), contact the Applied Biosystems
Care Center. To obtain a phone number for or to send an email to the center, go to
http://www.appliedbiosystems.com/support/contact.

Preface
Safety Conventions Used in This Document
xiii
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Safety Conventions Used in This Document
Safety Alert
Words
Four safety alert words appear in Applied Biosystems user documentation at points in
the document where you need to be aware of relevant hazards. Each alert
word—IMPORTANT, CAUTION, WARNING, DANGER—implies a particular
level of observation or action, as defined below.
Definitions
IMPORTANT! – Indicates information that is necessary for proper instrument operation,
accurate reagent kit use, or safe use of a chemical.
– Indicates a potentially hazardous situation that, if not avoided, may
result in minor or moderate injury. It may also be used to alert against unsafe practices.
– Indicates a potentially hazardous situation that, if not avoided,
could result in death or serious injury.
– Indicates an imminently hazardous situation that, if not avoided,
will result in death or serious injury. This signal word is limited to the most extreme
situations.
Except for IMPORTANTs, each safety alert word in an Applied Biosystems document
appears with an open triangle figure that contains a hazard symbol. These hazard
symbols are identical to the hazard symbols that are affixed to Applied Biosystems
instruments (see “Safety Symbols” on page xv).

Preface
Symbols on Instruments
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
xiv
Examples
IMPORTANT! You must create a separate sample entry spreadsheet for each 96-well
plate.
CHEMICAL HAZARD. TaqM an
®
Universal PCR Master Mix
may cause eye and skin irritation. Exposure may cause discomfort if swallowed or
inhaled. Read the MSDS, and follow the handling instructions. Wear appropriate
protective eyewear, clothing, and gloves.
PHYSICAL INJURY HAZARD. During instrument operation, the
the heated cover and sample block can reach temperatures in excess of 100 °C.
ELECTRICAL HAZARD. Grounding circuit continuity is vital for
the safe operation. Never operate the system with the grounding conductor disconnected.
Symbols on Instruments
Electrical
Symbols
The following table describes the electrical symbols that may be displayed on
Applied Biosystems instruments.
Symbol Description Symbol Description
Indicates the On position of the
main power switch.
Indicates a terminal that may be
connected to the signal ground
reference of another instrument.
This is not a protected ground
terminal.
Indicates the Off position of the
main power switch.
Indicates a protective grounding
terminal that must be connected
to earth ground before any other
electrical connections are made
to the instrument.
Indicates a standby switch by
which the instrument is switched
on to the Standby condition.
Hazardous voltage may be
present if this switch is on
standby.
Indicates a terminal that can
receive or supply alternating
current or voltage.
Indicates the On/Off position of a
push-push main power switch.
Indicates a terminal that can
receive or supply alternating or
direct current or voltage.

Preface
Symbols on Instruments
xv
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Safety Symbols
The following table describes the safety symbols that may be displayed on
Applied Biosystems instruments. Each symbol may appear by itself or with text that
explains the relevant hazard (see “Safety Labels on Instruments” on page xvi). These
safety symbols may also appear next to DANGERS, WARNINGS, and CAUTIONS that
occur in the text of this and other product-support documents.
Environmental
Symbols
The following symbol applies to all Applied Biosystems electrical and electronic
products placed on the European market after August 13, 2005.
Symbol Description Symbol Description
Indicates that you should consult
the manual for further information
and to proceed with appropriate
caution.
Indicates the presence of moving
parts and to proceed with
appropriate caution.
Indicates the presence of an
electrical shock hazard and to
proceed with appropriate caution.
Indicates the presence of a hot
surface or other high-temperature
hazard and to proceed with
appropriate caution.
Indicates the presence of a laser
inside the instrument and to
proceed with appropriate caution.
Symbol Description
Do not dispose of this product as unsorted municipal waste. Follow local
municipal waste ordinances for proper disposal provisions to reduce the
environmental impact of waste electrical and electronic equipment (WEEE).
European Union customers:
Call your local Applied Biosystems Customer Service office for equipment pick-up
and recycling. See http://www.appliedbiosystems.com for a list of customer
service offices in the European Union.

Preface
Safety Labels on Instruments
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
xvi
Safety Labels on Instruments
The following CAUTION, WARNING, and DANGER statements may be displayed on
Applied Biosystems instruments in combination with the safety symbols described in the
preceding section.
Locations of
Warnings
The StepOne and StepOnePlus instruments contain a warning at the location shown
below:
English Francais
CAUTION Hazardous chemicals. Read the
Material Safety Data Sheets (MSDSs) before
handling.
ATTENTION Produits chimiques
dangeureux. Lire les fiches techniques de
sûreté de matériels avant la manipulation des
produits.
CAUTION Hazardous waste. Refer to
MSDS(s) and local regulations for handling
and disposal.
ATTENTION Déchets dangereux. Lire les
fiches techniques de sûreté de matériels et la
régulation locale associées à la manipulation
et l'élimination des déchets.
CAUTION Hot surface. ATTENTION Surface brûlante.
DANGER High voltage. DANGER Haute tension.
WARNING To reduce the chance of electrical
shock, do not remove covers that require tool
access. No user-serviceable parts are inside.
Refer servicing to Applied Biosystems
qualified service personnel.
AVERTISSEMENT Pour éviter les risques
d'électrocution, ne pas retirer les capots dont
l'ouverture nécessite l'utilisation d'outils.
L’instrument ne contient aucune pièce
réparable par l’utilisateur. Toute intervention
doit être effectuée par le personnel de service
qualifié de Applied Biosystems.
CAUTION Moving parts. ATTENTION Parties mobiles.
DANGER Class 3B (III) visible and/or invisible
LED radiation present when open and
interlocks defeated. Avoid exposure to beam.
DANGER Rayonnement visible ou invisible
d’un faisceau LED de Classe 3B (III) en cas
d’ouverture et de neutralisation des
dispositifs de sécurité. Evitez toute
exposition au faisceau.

Preface
General Instrument Safety
xvii
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
General Instrument Safety
PHYSICAL INJURY HAZARD. Using the instrument in a manner
not specified by Applied Biosystems may result in personal injury or damage to the
instrument.
Moving and
Lifting the
Instrument
PHYSICAL INJURY HAZARD. The instrument is to be moved
and positioned only by the personnel or vendor specified in the applicable site
preparation guide. If you decide to lift or move the instrument after it has been installed,
do not attempt to lift or move the instrument without the assistance of others, the use of
appropriate moving equipment, and proper lifting techniques. Improper lifting can cause
painful and permanent back injury. Depending on the weight, moving or lifting an
instrument may require two or more persons.
Moving and
Lifting Computers
and Monitors
Do not attempt to lift or move the computer or the monitor without
the assistance of others. Depending on the weight of the computer and/or the monitor,
moving them may require two or more people.
Things to consider before lifting the computer and/or the monitor:
• Make sure that you have a secure, comfortable grip on the computer or the monitor
when lifting.
• Make sure that the path from where the object is to where it is being moved is clear
of obstructions.
• Do not lift an object and twist your torso at the same time.
• Keep your spine in a good neutral position while lifting with your legs.
• Participants should coordinate lift and move intentions with each other before
lifting and carrying.
• Instead of lifting the object from the packing box, carefully tilt the box on its side
and hold it stationary while someone slides the contents out of the box.
Operating the
Instrument
Ensure that anyone who operates the instrument has:
• Received instructions in both general safety practices for laboratories and specific
safety practices for the instrument.
• Read and understood all applicable Material Safety Data Sheets (MSDSs). See
“About MSDSs” on page xviii.
Cleaning or
Decontaminating
the Instrument
Before using a cleaning or decontamination method other than those
recommended by the manufacturer, verify with the manufacturer that the proposed
method will not damage the equipment.

Preface
Chemical Safety
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
xviii
Chemical Safety
Chemical Hazard
Warning
CHEMICAL HAZARD. Before handling any chemicals, refer to
the Material Safety Data Sheet (MSDS) provided by the manufacturer, and observe all
relevant precautions.
CHEMICAL STORAGE HAZARD. Never collect or store waste
in a glass container because of the risk of breaking or shattering. Reagent and waste
bottles can crack and leak. Each waste bottle should be secured in a low-density
polyethylene safety container with the cover fastened and the handles locked in the
upright position. Wear appropriate eyewear, clothing, and gloves when handling reagent
and waste bottles.
Chemical Safety
Guidelines
To minimize the hazards of chemicals:
• Read and understand the Material Safety Data Sheets (MSDSs) provided by the
chemical manufacturer before you store, handle, or work with any chemicals or
hazardous materials. (See “About MSDSs” on page xviii.)
• Minimize contact with chemicals. Wear appropriate personal protective equipment
when handling chemicals (for example, safety glasses, gloves, or protective
clothing). For more safety guidelines, consult the MSDS.
• Minimize the inhalation of chemicals. Do not leave chemical containers open. Use
only with adequate ventilation (for example, fume hood). For more safety
guidelines, consult the MSDS.
• Check regularly for chemical leaks or spills. If a leak or spill occurs, follow the
manufacturer’s cleanup procedures as recommended in the MSDS.
• Comply with all local, state/provincial, or national laws and regulations related to
chemical storage, handling, and disposal.
About MSDSs
Chemical manufacturers supply current Material Safety Data Sheets (MSDSs) with
shipments of hazardous chemicals to new customers. They also provide MSDSs with the
first shipment of a hazardous chemical to a customer after an MSDS has been updated.
MSDSs provide the safety information you need to store, handle, transport, and dispose
of the chemicals safely.
Each time you receive a new MSDS packaged with a hazardous chemical, be sure to
replace the appropriate MSDS in your files.
Obtaining
MSDSs
The MSDS for any chemical supplied by Applied Biosystems is available to you free
24 hours a day. To obtain MSDSs:
1. Go to https://docs.appliedbiosystems.com/msdssearch.html
2. In the Search field of the MSDS Search page:
a. Type in the chemical name, part number, or other information that you expect
to appear in the MSDS of interest.
b. Select the language of your choice.

Preface
Chemical Waste Safety
xix
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
c. Click Search.
3. To view, download, or print the document of interest:
a. Right-click the document title.
b. Select:
• Open – To view the document
• Save Target As – To download a PDF version of the document to a
destination that you choose
• Print Target – To print the document
4. To have a copy of an MSDS sent by fax or e-mail, in the Search Results page:
a. Select Fax or Email below the document title.
b. Click RETRIEVE DOCUMENTS at the end of the document list.
c. Enter the required information.
d. Click View/Deliver Selected Documents Now.
Note: For the MSDSs of chemicals not distributed by Applied Biosystems, contact the
chemical manufacturer.
Chemical Waste Safety
Chemical Waste
Hazard
HAZARDOUS WASTE. Refer to Material Safety Data Sheets and
local regulations for handling and disposal.
CHEMICAL WASTE HAZARD. Wastes produced by Applied
Biosystems instruments are potentially hazardous and can cause injury, illness, or death.
CHEMICAL STORAGE HAZARD. Never collect or store waste
in a glass container because of the risk of breaking or shattering. Reagent and waste
bottles can crack and leak. Each waste bottle should be secured in a low-density
polyethylene safety container with the cover fastened and the handles locked in the
upright position. Wear appropriate eyewear, clothing, and gloves when handling reagent
and waste bottles.
Chemical Waste
Safety Guidelines
To minimize the hazards of chemical waste:
• Read and understand the Material Safety Data Sheets (MSDSs) provided by the
manufacturers of the chemicals in the waste container before you store, handle, or
dispose of chemical waste.

Preface
Electrical Safety
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
xx
• Provide primary and secondary waste containers. (A primary waste container holds
the immediate waste. A secondary container contains spills or leaks from the
primary container. Both containers must be compatible with the waste material and
meet federal, state, and local requirements for container storage.)
• Minimize contact with chemicals. Wear appropriate personal protective equipment
when handling chemicals (for example, safety glasses, gloves, or protective
clothing). For more safety guidelines, consult the MSDS.
• Minimize the inhalation of chemicals. Do not leave chemical containers open. Use
only with adequate ventilation (for example, fume hood). For more safety
guidelines, consult the MSDS.
• Handle chemical wastes in a fume hood.
• After emptying a waste container, seal it with the cap provided.
• Dispose of the contents of the waste tray and waste bottle in accordance with good
laboratory practices and local, state/provincial, or national environmental and
health regulations.
Waste Disposal
If potentially hazardous waste is generated when you operate the instrument, you must:
• Characterize (by analysis if necessary) the waste generated by the particular
applications, reagents, and substrates used in your laboratory.
• Ensure the health and safety of all personnel in your laboratory.
• Ensure that the instrument waste is stored, transferred, transported, and disposed of
according to all local, state/provincial, and/or national regulations.
IMPORTANT! Radioactive or biohazardous materials may require special handling, and
disposal limitations may apply.
Electrical Safety
ELECTRICAL SHOCK HAZARD. Severe electrical shock can
result from operating the StepOne or StepOnePlus instrument without its instrument
panels in place. Do not remove instrument panels. High-voltage contacts are exposed
when instrument panels are removed from the instrument.
Fuses
FIRE HAZARD. Improper fuses or high-voltage supply can
damage the instrument wiring system and cause a fire. Before turning on the instrument,
verify that the fuses are properly installed and that the instrument voltage matches the
power supply in your laboratory.
FIRE HAZARD. For continued protection against the risk of fire,
replace fuses only with fuses of the type and rating specified for the instrument.

Preface
LED Safety
xxi
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Power
ELECTRICAL HAZARD. Grounding circuit continuity is vital for
the safe operation of equipment. Never operate equipment with the grounding conductor
disconnected.
ELECTRICAL HAZARD. Use properly configured and approved
line cords for the voltage supply in your facility.
ELECTRICAL HAZARD. Plug the instrument into a properly
grounded receptacle with adequate current capacity.
Overvoltage
Rating
The StepOne and StepOnePlus instruments have an installation (overvoltage) category
of II, and they are classified as portable equipment.
LED Safety
To ensure safe LED operation:
• The system must be maintained by an Applied Biosystems Technical
Representative.
• All instrument panels must be in place on the instrument while the instrument is
operating. When all panels are installed, there is no detectable radiation present. If
any panel is removed when the LED is operating (during service with safety
interlocks disabled), you may be exposed to LED emissions in excess of the
Class 3B rating.
• Do not remove safety labels or disable safety interlocks.
Biological Hazard Safety
General
Biohazard
BIOHAZARD. Biological samples such as tissues, body fluids,
infectious agents, and blood of humans and other animals have the potential to transmit
infectious diseases. Follow all applicable local, state/provincial, and/or national
regulations. Wear appropriate protective equipment, which includes but is not limited to:
protective eyewear, face shield, clothing/lab coat, and gloves. All work should be
performed in properly equipped facilities using the appropriate safety equipment (for
example, physical containment devices). Individuals should be trained according to
applicable regulatory and company/institution requirements before working with
potentially infectious materials. Read and follow the applicable guidelines and/or
regulatory requirements in the following:
• U.S. Department of Health and Human Services guidelines published in Biosafety
in Microbiological and Biomedical Laboratories (stock no. 017-040-00547-4;
http://bmbl.od.nih.gov)
• Occupational Safety and Health Standards, Bloodborne Pathogens (29
CFR§1910.1030; http://www.access.gpo.gov/ nara/cfr/waisidx_01/
29cfr1910a_01.html).

Preface
Workstation Safety
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
xxii
• Your company’s/institution’s Biosafety Program protocols for working
with/handling potentially infectious materials.
More information about biohazard guidelines is available at:
http://www.cdc.gov
Workstation Safety
Correct ergonomic configuration of your workstation can reduce or prevent effects such
as fatigue, pain, and strain. Minimize or eliminate these effects by configuring your
workstation to promote neutral or relaxed working positions.
MUSCULOSKELETAL AND REPETITIVE MOTION
HAZARD. These hazards are caused by potential risk factors that include but are not
limited to repetitive motion, awkward posture, forceful exertion, holding static unhealthy
positions, contact pressure, and other workstation environmental factors.
To minimize musculoskeletal and repetitive motion risks:
• Use equipment that comfortably supports you in neutral working positions and
allows adequate accessibility to the keyboard, monitor, and mouse.
• Position the keyboard, mouse, and monitor to promote relaxed body and head
postures.
Safety and Electromagnetic Compatibility (EMC) Standards
U.S. and
Canadian Safety
Standards
The StepOne and StepOnePlus instruments have been tested to and comply with
standard:
UL 61010A-1/CAN/CSA C22.2 No. 1010.1-92, “Safety Requirements for Electrical
Equipment for Measurement, Control, and Laboratory Use, Part 1: General
Requirements.”
UL 61010A-2-010/CAN/CSA 1010.2.010, “Particular Requirements for Laboratory
Equipment for the Heating of Materials.”
FDA “Radiation Control for Health and Safety Act of 1968 Performance Standard 21
CFR 1040.10 and 1040.11,” as applicable.
Canadian EMC
Standard
This instrument has been tested to and complies with ICES-001, Issue 3: “Industrial,
Scientific, and Medical Radio Frequency Generators.”
European Safety
and EMC
Standards
Safety
This instrument meets European requirements for safety (Low Voltage Directive
73/23/EEC). This instrument has been tested to and complies with standards EN 61010-
1:2001, “Safety Requirements for Electrical Equipment for Measurement, Control and
Laboratory Use, Part 1: General Requirements.”

Preface
Safety and Electromagnetic Compatibility (EMC) Standards
xxiii
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
EN 61010-2-010, “Particular Requirements for Laboratory Equipment for the Heating of
Materials.”
EN 61010-2-081, “Particular Requirements for Automatic and Semi-Automatic
Laboratory Equipment for Analysis and Other Purposes.”
EN 60825-1, “Radiation Safety of Laser Products, Equipment Classification,
Requirements, and User’s Guide.
EMC
This instrument meets European requirements for emission and immunity (EMC
Directive 89/336/EEC). This instrument has been tested to and complies with standard
EN 61326 (Group 1, Class B), “Electrical Equipment for Measurement, Control and
Laboratory Use – EMC Requirements.”
Australian EMC
Standards
This instrument has been tested to and complies with standard AS/NZS 2064, “Limits
and Methods Measurement of Electromagnetic Disturbance Characteristics of Industrial,
Scientific, and Medical (ISM) Radio-frequency Equipment.”
Preface
Safety and Electromagnetic Compatibility (EMC) Standards
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
xxiv

Chapter 1
1
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Get Started
This chapter covers:
■ About the StepOne
™
and StepOnePlus
™
Systems . . . . . . . . . . . . . . . . . . . . . . . . . . 2
■ Supported Consumables . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 4
■ About Standard Curve Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
■ How to Use This Guide. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
■ About the Example Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
■ Example Experiment Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12
Note: For more information about any of the topics discussed in this guide, access the
Help from within Applied Biosystems StepOne
™
Real-Time PCR Software v2.0 by
pressing F1, clicking in the toolbar, or selecting HelpStepOne Software Help.

Chapter 1 Get Started
About the StepOne
™
and StepOnePlus
™
Systems
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
2
Notes
About the StepOne
™
and StepOnePlus
™
Systems
There are two models available for this Real-Time PCR System:
The StepOne and StepOnePlus systems use fluorescent-based polymerase chain reaction
(PCR) reagents to provide:
• Quantitative detection of target nucleic acid sequences (targets) using real-time
analysis.
• Qualitative detection of targets using post-PCR (endpoint) analysis.
• Qualitative analysis of the PCR product (achieved by melt curve analysis that occurs
post-PCR).
About Data
Collection
The StepOne and StepOnePlus systems collect raw fluorescence data at different points
during a PCR, depending on the type of run that the instruments perform:
Regardless of the run type, a data collection point or read on the StepOne
™
or
StepOnePlus
™
instrument consists of three phases:
1. Excitation – The instrument illuminates all wells of the reaction plate within the
instrument, exciting the fluorophores in each reaction.
System Features
Applied Biosystems StepOne
™
Real-Time
PCR System (StepOne™ system)
• 48-well platform
• Three-color system
Applied Biosystems StepOnePlus
™
Real-
Time PCR System (StepOnePlus
™
system)
• 96-well platform
• Four-color system
• VeriFlex
™
sample blocks
Run Type Data Collection Point
Real-time runs Standard curve The instrument collects data following each
extension step of the PCR.
Relative standard
curve
Comparative C
T
(ΔΔC
T
)
Post-PCR
(endpoint) runs
Genotyping The instrument collects data:
• Before the PCR (For presence/absence
experiments, data collection before the PCR is
optional, but recommended.)
• (Optional) During the PCR. The instrument can
collect data during the run (real-time); collecting
data during the run can be helpful for
troubleshooting.
•After the PCR
Presence/absence

Chapter 1 Get Started
About the StepOne
™
and StepOnePlus
™
Systems
3
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
2. Emission – The instrument optics collect the residual fluorescence emitted from the
wells of the reaction plate. The resulting image collected by the device consists only
of light that corresponds to the range of emission wavelengths.
3. Collection – The instrument assembles a digital representation of the residual
fluorescence collected over a fixed time interval. The StepOne
™
software stores the
raw fluorescent image for analysis.
After a run, the StepOne software uses calibration data (spatial, dye, and background) to
determine the location and intensity of the fluorescent signals in each read, the dye
associated with each fluorescent signal, and the significance of the signal.
About the Filters
The StepOne and StepOnePlus systems use the following filters:
About the
VeriFlex
™
Te c h no l o gy
The StepOnePlus instrument contains six independently thermally regulated VeriFlex
™
blocks to help you optimize your thermal cycling conditions. You can set a different
temperature for one or more of the VeriFlex blocks, creating up to six different zones for
samples, or you can set the same temperature for each of the VeriFlex blocks.
For More
Information
For information on:
• The StepOne and StepOnePlus systems, refer to Applied Biosystems StepOne
™
Real-Time PCR Software Help.
Note: To access the Help, select HelpStepOne Software Help from within the
StepOne software.
• Genotyping experiments, refer to Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems Getting Started Guide for Genotyping Experiments.
• Presence/absence experiments, refer to Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems Getting Started Guide for Presence/Absence
Experiments.
• Relative standard curve and/or comparative C
T
(ΔΔC
T
) experiments, refer to
Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems Getting
Started Guide for Relative Standard Curve and Comparative C
T
Experiments.
StepOne system StepOnePlus system
Filter Dye Filter Dye
1FAM
™
dye 1 FAM
™
dye
SYBR
®
Green dye SYBR
®
Green dye
2JOE
™
dye 2 JOE
™
dye
VIC
®
dye VIC
®
dye
3ROX
™
dye 3 TAMRA
™
dye
NED
™
dye
4ROX
™
dye

Chapter 1 Get Started
Supported Consumables
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
4
Notes
Supported Consumables
StepOne System
The StepOne system supports the consumables listed below. These consumables are for
use with both standard and Fast reagents/protocols.
IMPORTANT! Use only Fast consumables (reaction plates, tube strips, and tubes) with
the StepOne and StepOnePlus systems, even when performing an experiment with
standard reagents.
Consumable Part Number
•MicroAmp
™
Fast Optical 48-Well Reaction Plate
•MicroAmp
™
48-Well Optical Adhesive Film
• 4375816
• 4375323 and
4375928
•MicroAmp
™
Fast 8-Tube Strip
•MicroAmp
™
Optical 8-Cap Strip
• 4358293
• 4323032
•MicroAmp
®
Fast Reaction Tube with Cap • 4358297
•MicroAmp
™
Fast 48-Well Tray
•MicroAmp
™
48-Well Base Adaptor
•MicroAmp
™
96-Well Support Base
• 4375282
• 4375284
• 4379590
G
A
C
D
C
F
B
C
# Consumable
AMicroAmp
™
Fast Optical 48-Well Reaction Plate
BMicroAmp
™
Fast 48-Well Tray
CMicroAmp
™
96-Well Support Base
DMicroAmp
™
Optical 8-Cap Strip
EMicroAmp
™
Fast 8-Tube Strip
FMicroAmp
®
Fast Reaction Tube with Cap
GMicroAmp
™
48-Well Optical Adhesive Film
HMicroAmp
™
48-Well Base Adaptor
E
B
H
H

Chapter 1 Get Started
Supported Consumables
5
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
StepOnePlus
System
The StepOnePlus system supports the consumables listed below. These consumables are
for use with both standard and Fast reagents/protocols.
IMPORTANT! Use only Fast consumables (reaction plates, tube strips, and tubes) with
the StepOne and StepOnePlus systems, even when performing an experiment with
standard reagents.
Consumable Part Number
•MicroAmp
™
Fast Optical 96-Well Reaction Plate with
Barcode
•MicroAmp
™
Optical Adhesive Film
• 4346906 and
4366932
• 4360954 and
4311971
•MicroAmp
™
Fast 8-Tube Strip
•MicroAmp
™
Optical 8-Cap Strip
• 4358293
• 4323032
•MicroAmp
®
Fast Reaction Tube with Cap • 4358297
•MicroAmp
™
96-Well Tray for VeriFlex
™
Blocks
•MicroAmp
™
96-Well Support Base
• 4379983
• 4379590
•MicroAmp
™
Adhesive Film Applicator
•MicroAmp
™
Cap Installing Tool (Handle)
• 4333183
• 4330015
G
A
C
F
B
C
# Consumable
AMicroAmp
™
Fast Optical 96-Well Reaction Plate
BMicroAmp
™
96-Well Tray for VeriFlex
™
Blocks
CMicroAmp
™
96-Well Support Base
DMicroAmp
™
Optical 8-Cap Strip
EMicroAmp
™
Fast 8-Tube Strip
FMicroAmp
®
Fast Reaction Tube with Cap
GMicroAmp
™
Optical Adhesive Film
D
C
E
B

Chapter 1 Get Started
About Standard Curve Experiments
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
6
Notes
About Standard Curve Experiments
Real-Time PCR
Experiments
Standard curve experiments are real-time PCR experiments. In real-time PCR
experiments:
• The instrument monitors the progress of the PCR as it occurs (Kwok and Higuchi,
1989).
• Data are collected throughout the PCR process.
• Reactions are characterized by the point in time during cycling when amplification
of a target is first detected (Saiki et al., 1985).
Note: In this guide, the term experiment refers to the entire process of performing a run
using the StepOne or StepOnePlus system, including setup, run and analysis.
About Standard
Curve
Experiments
The standard curve method is used to determine the absolute target quantity in samples.
With the standard curve method, the StepOne software measures amplification of the
target in samples and in a standard dilution series. Data from the standard dilution series
are used to generate the standard curve. Using the standard curve, the software
interpolates the absolute quantity of target in the samples.
Components
The following components are required when setting up PCR reactions for standard
curve experiments:
• Sample – The sample in which the quantity of the target is unknown.
• Standard – A sample that contains known standard quantities; used in quantitation
experiments to generate standard curves.
• Standard dilution series – A set of standards containing a range of known
quantities. The standard dilution series is prepared by serially diluting standards.
• Replicates – The total number of identical reactions containing identical samples,
components, and volumes.
• Negative Controls – Wells that contain water or buffer instead of sample template.
No amplification of the target should occur in negative control wells.
PCR Options
When performing real-time PCR, choose between:
• Singleplex and multiplex PCR (below)
and
• 1-step and 2-step RT-PCR (page 7)
Singleplex vs. Multiplex PCR
You can perform a PCR reaction using either:
• Singleplex PCR – In singleplex PCR a single primer set is present in the reaction
tube or well. Only one target or endogenous control can be amplified per reaction.
or

Chapter 1 Get Started
About Standard Curve Experiments
7
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
• Multiplex PCR – In multiplex PCR, two or more primer sets are present in the
reaction tube or well. Each set amplifies a specific target or endogenous control.
Typically, a probe labeled with FAM
™
dye detects the target and a probe labeled
with VIC
®
dye detects the endogenous control.
IMPORTANT! SYBR
®
Green reagents cannot be used for multiplex PCR.
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA
™
dye
as a reporter or quencher with the StepOne system. TAMRA dye may be used as a
reporter or quencher with the StepOnePlus system.
1- vs. 2-Step RT-PCR
You can perform reverse transcription (RT) and PCR in a single reaction (1-step) or in
separate reactions (2-step). The reagent configuration you use depends on whether you
are performing 1- or 2-step RT-PCR:
• In 1-step RT-PCR, RT and PCR take place in one buffer system, which provides the
convenience of a single-tube preparation for RT and PCR amplification. However,
you cannot use Fast PCR master mix or the carryover prevention enzyme,
AmpErase
®
UNG (uracil-N-glycosylase), to perform 1-step RT-PCR.
• 2-step RT-PCR is performed in two separate reactions: First, total RNA is reverse-
transcribed into cDNA, then the cDNA is amplified by PCR. This method is useful
for detecting multiple transcripts from a single cDNA template or for storing cDNA
aliquots for later use. The AmpErase
®
UNG enzyme can be used to prevent
carryover contamination.
Note: For more information on AmpErase
®
UNG, refer to the Applied Biosystems
StepOne
™
and StepOnePlus
™
Real-Time PCR Systems Reagent Guide.
GR2331
Target Primer Set
Endogenous Control
Primer Set
cDNA
Singleplex PCR
Multiplex PCR

Chapter 1 Get Started
About Standard Curve Experiments
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
8
Notes
Supported
Reagents
TaqM an
®
and SYBR
®
Green Reagents
Applied Biosystems offers TaqMan
®
and SYBR
®
Green reagents for use on the StepOne
and StepOnePlus systems. Both reagent types are briefly described in the table below.
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA dye as a
reporter or quencher with the StepOne system. TAMRA dye may be used as a reporter or
quencher with the StepOnePlus system.
Reagent Type Process
Ta q Ma n
®
reagents or kits
Description
TaqMan reagents use a fluorogenic probe to
enable detection of a specific PCR product as
it accumulates during PCR cycles.
Advantages
• Increased specificity with the addition of a
fluorogenic probe.
• Provides multiplex capability.
• Preformulated assays, optimized to run
under universal thermal cycling conditions,
are available.
• Can be used for either 1- or 2-step RT-PCR.
Limitations
Requires synthesis of a unique fluorogenic
probe.
SYBR
®
Green reagents
Description
SYBR Green reagents use SYBR
®
Green I dye,
a double-stranded DNA binding dye, to detect
PCR products as they accumulate during PCR
cycles.
Advantages
• Economical (no probe needed).
• Allows for melt curve analysis to measure
the Tm of all PCR products.
• Can be used for either 1- or 2-step RT-PCR.
Limitations
Binds nonspecifically to all double-stranded
DNA sequences. To avoid false positive
signals, check for nonspecific product
formation using melt curve or gel analysis.
b. Denatured Template and Annealing of Assay Components
Forward primer
Reverse primer
Q
MGB
F
LEGEND
FAM™ dye
Quencher
Minor Groove
Binder
AmpliTaq Gold
®
DNA Polymerase
Probe
Primer
Template
Extended Primer
5'3'
a. Assay Components
cDNA Template
Forward primer
Reverse primer
Probe
Q
MGB
F
Probe
Q
MGB
F
5'3'
PCR and Detection of cDNA
cDNA
5'3'
5'3'
c. Signal Generation
Reverse primer
F
Forward primer
5'3'
3'5'
5'3'
MGB
Q
FORWARD
PRIMER
REVERSE
PRIMER
Step 1: Reaction setup
The SYBR
®
Green I dye
fluoresces when bound to
double-stranded DNA.
Step 2: Denaturation
When the DNA is denatured into
single-stranded DNA, the
®
SYBR Green I dye is released and
the fluorescence is drastically reduced.
Step 3: Polymerization
During extension, primers
anneal and PCR product
is generated.
Step 4: Polymerization completed
SYBR
®
Green I dye binds to the
double-stranded product,
resulting in a net increase in
fluorescence detected by the
instrument.

Chapter 1 Get Started
How to Use This Guide
9
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Other Reagents
You can use other fluorescent-based reagents on the StepOne and StepOnePlus systems,
but note the following:
• You must design your experiment using Advanced Setup instead of the Design
Wizard. (See “Advanced Setup Workflow” on page 116.)
• For Applied Biosystems TaqMan and SYBR Green reagents, the StepOne software
automatically calculates reaction volumes in the Reaction Setup screen.
For More
Information
For more information on real-time PCR experiments, PCR options, and reagents, refer to
the Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems Reagent
Guide.
How to Use This Guide
This guide functions as both a tutorial and as a guide for performing your own
experiments.
Using This Guide
as a Tutorial
By using the example experiment data provided with the StepOne software, you can use
this guide as a tutorial for performing a standard curve experiment on a StepOne or
StepOnePlus system. Follow the procedures in Chapters 2 to 5:
For more information, see “About the Example Experiment” on page 10.
Using This Guide
With Your Own
Experiments
After completing the tutorial exercises in Chapters 2 to 5, you can use this guide to lead
you through your own standard curve experiments. Each procedure in Chapters 2 to 5
includes a set of guidelines that you can use to perform your own experiments.
Chapter Procedure
2 Design the experiment using the Design Wizard in the StepOne software.
3 Prepare the experiment, using the reagents and volumes calculated by the
Design Wizard in Chapter 2.
4 Run the experiment on a StepOne or StepOnePlus instrument (standalone or
colocated layout)
5 Analyze the results.

Chapter 1 Get Started
About the Example Experiment
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
10
Notes
Additionally, you can use one of the other workflows provided in the StepOne software to
perform your experiments. The table below provides a summary of all the workflows
available in the StepOne software.
About the Example Experiment
To illustrate how to perform standard curve experiments, this guide leads you through the
process of designing, preparing, running, and analyzing an example experiment. The
example experiment represents a typical setup that you can use to quickly familiarize
yourself with a StepOne or StepOnePlus system.
Description
The objective of the standard curve example experiment is to determine the quantity of
the RNase P gene in two populations.
In the standard curve example experiment:
• The samples are genomic DNA isolated from two populations.
• The target is the RNase P gene.
• One standard curve is set up for the RNase P gene (target). The standard used for the
standard dilution series contains known quantities of the RNase P gene. Because a
single target is being studied, only one standard curve is required.
Note: In experiments where multiple targets are being studied, a standard curve is
required for each target.
• Three replicates of each sample and each dilution point in the standard curve are
performed to ensure statistical significance.
Workflow Description See...
Design Wizard Set up a new experiment with guidance from the software.
The Design Wizard walks you through best practices as
you create your own experiment. The Design Wizard is
recommended for new users.
Note: Design options are more limited in the Design
Wizard than in Advanced Setup.
Chapter 2
Advanced
Setup
Set up a new experiment using advanced options.
Advanced Setup allows design flexibility as you create
your own experiment. Advanced Setup is recommended
for experienced users.
page 116
QuickStart Run a new experiment with no plate setup information. If
desired, you can add all design parameters after the run.
page 117
Template Set up a new experiment using setup information from a
template.
page 119
Export/Import Import experiment designs from ASCII text files that
contain experiment setup information.
page 121

Chapter 1 Get Started
About the Example Experiment
11
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
• The experiment is designed for singleplex PCR, where every well contains a
primer/probe set for a single target.
• Reactions are set up for 2-step RT-PCR.
• Primer/probe sets are from Applied Biosystems RNase P assay.
Note: The human RNase P FAM
™
dye-labeled MGB probe is not available as a
TaqMan
®
Gene Expression Assay. It can be ordered as a Custom TaqMan
®
Gene
Expression Assay (PN 4331348).
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA dye as
a reporter or quencher with the StepOne system. TAMRA dye may be used as a
reporter or quencher with the StepOnePlus system.
Reaction Plate
Layout
The standard curve example experiment was created for a StepOne instrument. For the
StepOne instrument, the software displays a 48-well reaction plate layout:
You can create the example experiment for a StepOnePlus instrument; however, your
reaction plate layout will differ from the 48-well reaction plate layout shown throughout
this guide. For the StepOnePlus instrument, the software displays a 96-well reaction plate
layout:

Chapter 1 Get Started
Example Experiment Workflow
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
12
Notes
About the
Example
Experiment Data
In this getting started guide you will use two files:
• In Chapter 2, you will create a standard curve example experiment file that contains
setup data and save it to the experiments folder on your computer:
<drive>:\Applied Biosystems\<software name>\experiments\
Standard Curve Example.eds
• In Chapter 5, you will view results in a standard curve example experiment file that
contains run data. The data file for the example experiment installs with the
StepOne software. You can find the data file for the example experiment on your
computer:
<drive>:\Applied Biosystems\<software name>\experiments\examples\
Standard Curve Example.eds
where:
• <drive> is the computer hard drive on which the StepOne software is installed. The
default installation drive for the software is the D drive.
• <software name> is the current version of the StepOne software.
Data Files in the Examples Folder
The examples folder contains several data files that you can reference when analyzing
your own data, as listed below. The data files install with the StepOne software.
Note: Be sure to use the Standard Curve Example.eds file when performing the tutorial
procedures in this guide. The 96-Well Standard Curve Example.eds file is a different
example of the standard curve method.
Example Experiment Workflow
The figure on page 13 shows the workflow for the standard curve example experiment.
StepOne Instrument StepOnePlus Instrument
Comparative CT Example.eds 96-Well Comparative CT Example.eds
Multiplex Example.eds 96-Well Multiplex Example.eds
Genotyping Example.eds 96-Well Genotyping Example.eds
Presence Absence Example.eds 96-Well Presence Absence Example.eds
Relative Standard Curve Example.eds 96-Well Relative Standard Curve
Example.eds
RNase P Experiment.eds 96-Well RNase P Experiment.eds
Standard Curve Example.eds 96-Well Standard Curve Example.eds
SYBR Example.eds 96-Well SYBR Example.eds

Chapter 1 Get Started
Example Experiment Workflow
13
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Design the Experiment (Chapter 2)
1. Create a new experiment.
2. Define the experiment properties.
3. Define the methods and materials.
4. Set up the targets.
5. Set up the standards.
6. Set up the samples.
7. Set up the run method.
8. Review the reaction setup.
9. Order materials for the experiment.
10.Finish the Design Wizard.
Prepare the Reactions (Chapter 3)
1. Prepare the sample dilutions.
2. Prepare the standard dilution series.
3. Prepare the reaction mix for each target assay.
4. Prepare the reaction plate.
Run the Experiment (Chapter 4)
1. Prepare for the run.
2. (Optional) Enable the notification settings.
3. Start the run.
4. Monitor the run.
5. Unload the instrument and transfer the data.
Start Experiment
End Experiment
Analyze the Experiment (Chapter 5)
Section 1, Review Results:
1. Analyze.
2. View the standard curve.
3. View the amplification plot.
4. View the results in a table.
5. Publish the data.
Section 2, Troubleshoot (If Needed):
1. View the analysis settings; adjust the baseline/threshold.
2. View the quality summary.
3. Omit wells.
4. View the multicomponent plot.
5. View the raw data plot.

Chapter 1 Get Started
Example Experiment Workflow
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
14
Notes

Chapter 2
15
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Design the Experiment
This chapter covers:
■ Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16
■ Create a New Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
■ Define the Experiment Properties . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20
■ Define the Methods and Materials . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22
■ Set Up the Targets. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25
■ Set Up the Standards. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27
■ Set Up the Samples. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29
■ Set Up the Run Method . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 31
■ Review the Reaction Setup . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33
■ Order Materials for the Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 38
■ Finish the Design Wizard . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 42
Note: For more information about any of the topics discussed in this guide, access the
Help from within Applied Biosystems StepOne
™
Real-Time PCR Software v2.0 by
pressing F1, clicking in the toolbar, or selecting HelpStepOne Software Help.

Chapter 2 Design the Experiment
Chapter Overview
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
16
Notes
Chapter Overview
This chapter explains how to use the Design Wizard in the StepOne
™
software to set up
the standard curve example experiment. The Design Wizard walks you through Applied
Biosystems recommended best practices as you enter design parameters for the example
experiment.
Example
Experiment
Workflow
The workflow for designing the example experiment provided with this getting started
guide is shown below.
Note: Design the example experiment using the Design Wizard in the StepOne software.
When you design your own experiments, you can select alternate workflows (see “Using
This Guide With Your Own Experiments” on page 9).
Analyze the Experiment (Chapter 5)
Start Experiment
Prepare the Reactions (Chapter 3)
Run the Experiment (Chapter 4)
End Experiment
Design the Experiment (Chapter 2)
1. Create a new experiment.
2. Define the experiment properties.
3. Define the methods and materials.
4. Set up the targets.
5. Set up the standards.
6. Set up the samples.
7. Set up the run method.
8. Review the reaction setup.
9. Order materials for the experiment.
10. Finish the Design Wizard.

Chapter 2 Design the Experiment
Create a New Experiment
17
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Create a New Experiment
Create a new experiment using the Design Wizard in the StepOne software.
Create an
Experiment
1. Double-click (StepOne software shortcut) or select
StartAll ProgramsApplied BiosystemsStepOne Software
<software name>
where <software name> is the current version of the StepOne software.
2. From the Home screen, click Design Wizard to open the Design Wizard.
3. See “Software Elements” on page 18 for information on navigating within the
Design Wizard.
2
Chapter 2 Design the Experiment
Create a New Experiment
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
18
Notes
Software
Elements
The StepOne software elements for the Design Wizard are illustrated below.
1. Menu bar – Displays the menus available in the software:
•File
•Edit
• Instrument
•Analysis
• Tools
•Help
2. Toolbar – Displays the tools available in the software:
• New Experiment
•Open
•Close
• Send Experiment to Instrument
• Download Experiment from Instrument
3. Experiment header - Displays the experiment name, experiment type, and reagents
for the open experiment.
4. Navigation pane – Provides links to all screens in the Design Wizard:
• Experiment Properties
• Methods & Materials
•Targets
•Standards
• Samples
•Run Method
• Reaction Setup
• Materials List
Note: The Design Wizard initially displays the Quantitation - Standard Curve
experiment type. The available Design Wizard screens may change when you select
a different experiment type. For example, the Relative Quantitation Settings screen
is not displayed until you select the relative standard curve or comparative C
T
(ΔΔC
T
) experiment type.
5. Experiment tab(s) – Displays a tab for each open experiment.

Chapter 2 Design the Experiment
Create a New Experiment
19
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
1
2
3
4
5

Chapter 2 Design the Experiment
Define the Experiment Properties
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
20
Notes
Define the Experiment Properties
On the Experiment Properties screen, enter identifying information for the experiment,
select the instrument type, then select the type of experiment to design.
About the
Example
Experiment
In the standard curve example experiment:
• The experiment is identified as an example.
• The instrument selected to run the experiment is the StepOne instrument.
•A MicroAmp
™
Fast Optical 48-Well Reaction Plate is used.
• The experiment type is quantitation.
Complete the
Experiment
Properties
Screen
1. Click the Experiment Name field, then enter Standard Curve Example.
Note: The experiment header updates with the experiment name you entered.
2. Leave the Barcode field empty.
Note: The MicroAmp Fast Optical 48-Well Reaction Plate does not have a barcode.
3. Click the User Name field, then enter Example User.
4. Click the Comments field, then enter Standard Curve Getting Started Guide
Example.
5. Select StepOne
™
Instrument (48 Wells).
Note: The example experiment was created for a StepOne instrument. You can
create the example experiment for a StepOnePlus instrument; however, your
reaction plate layout will differ from the layout shown in this guide. The software
displays a 48-well reaction plate layout for the StepOne instrument and a 96-well
reaction plate layout for the StepOnePlus instrument. To create the example
experiment for a StepOnePlus instrument, select StepOnePlus
™
Instrument
(96 Wells).
6. Select Quantitation for the experiment type.
7. Click Next >.
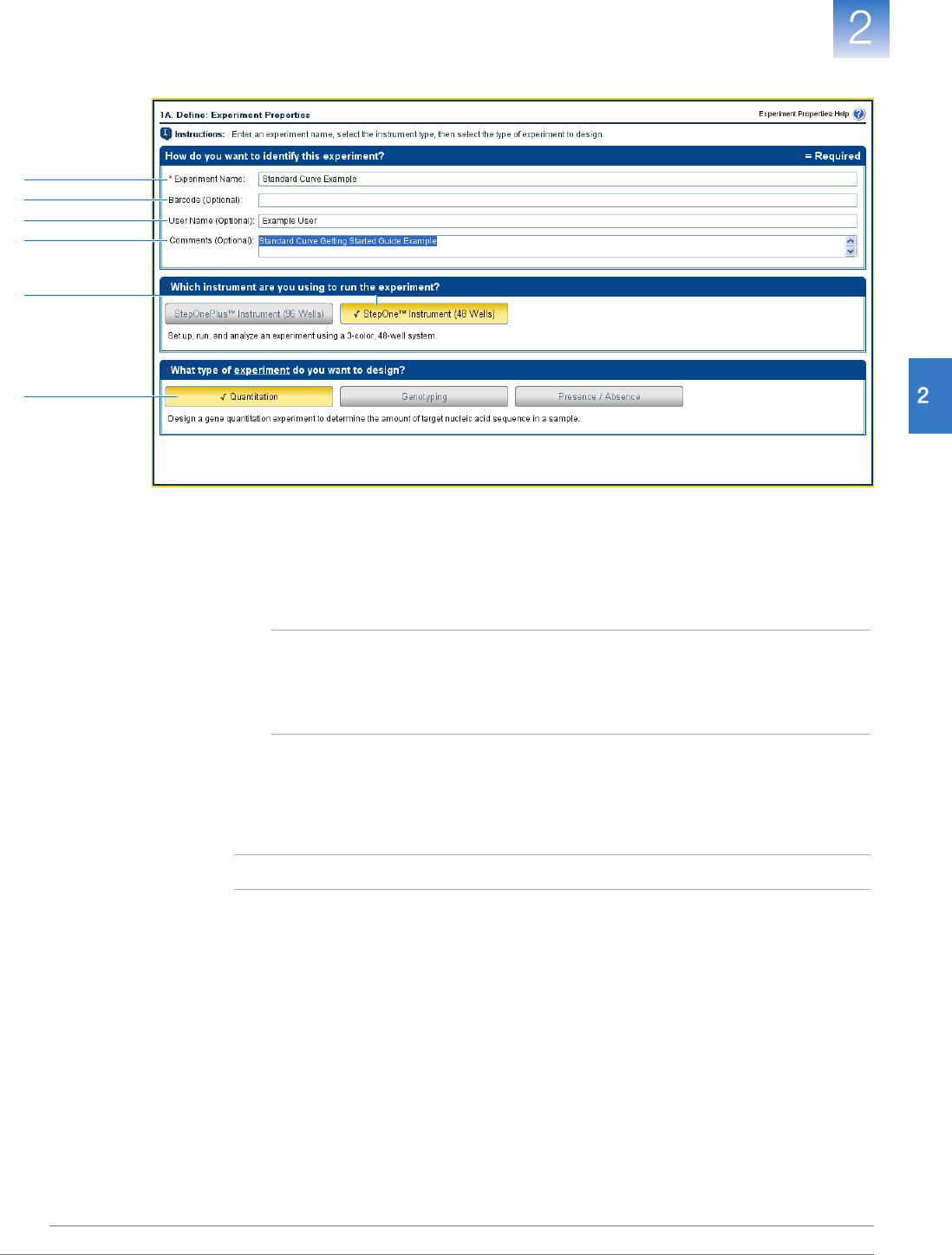
Chapter 2 Design the Experiment
Define the Experiment Properties
21
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Design
Guidelines
When you design your own standard curve experiment:
• Enter an experiment name:
– Enter a name that is descriptive and easy to remember. You can enter up to 100
characters in the Experiment Name field.
Note: You cannot use the following characters in the Experiment Name field:
forward slash (/), backslash (\), greater than sign (>), less than sign (<), asterisk
(*), question mark (?), quotation mark ("), vertical line (|), colon (:), or
semicolon (;).
– The experiment name is used as the default file name.
• (Optional) If you are using a MicroAmp
™
Fast Optical 96-Well Reaction Plate,
enter a barcode to identify the barcode on the reaction plate. You can enter up to 100
characters in the Barcode field.
Note: The MicroAmp Fast Optical 48-Well Reaction Plate does not have a barcode.
• (Optional) Enter a user name to identify the owner of the experiment. You can enter
up to 100 characters in the User Name field.
• (Optional) Enter comments to describe the experiment. You can enter up to 1000
characters in the Comments field.
1
2
3
4
6
5

Chapter 2 Design the Experiment
Define the Methods and Materials
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
22
Notes
• Select the instrument you are using to run the experiment:
– StepOne
™
Instrument (48 Wells)
– StepOnePlus
™
Instrument (96 Wells)
Note: You can use StepOne Software v2.0 or later to design experiments for both
the StepOne and StepOnePlus instruments. The instrument you select in the
Experiment Properties screen affects the reaction plate layout and materials list.
Note: To set the default instrument type, select ToolsPreferences, then select the
General tab (default). From the Default Instrument Type dropdown menu, select
the appropriate instrument.
• Select Quantitation as the experiment type.
For More
Information
For more information on:
• Completing the Experiment Properties screen, access the StepOne Software Help by
clicking or pressing F1.
• Consumables, see “Supported Consumables” on page 4.
• Quantitation experiments, refer to the Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems Reagent Guide.
Define the Methods and Materials
On the Methods & Materials screen, select the quantitation method, reagents, ramp
speed, and PCR template to use for the experiment.
About the
Example
Experiment
In the standard curve example experiment:
• The standard curve quantitation method is used.
•TaqMan
®
reagents are used.
• The standard ramp speed is used in the instrument run.
• Purified gDNA (isolated from two populations) is the template type. Before using
gDNA template, you must first extract the gDNA from tissue or sample.
Complete the
Methods &
Materials Screen
1. Select Standard Curve as the quantitation method.
2. Select TaqMan
®
Reagents for the reagents.
3. Select Standard (~ 2 hours to complete a run) for the ramp speed.
4. Select gDNA (genomic DNA) for the template type.
5. Click Next >.

Chapter 2 Design the Experiment
Define the Methods and Materials
23
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Design
Guidelines
When you design your own standard curve experiment:
• Select Standard Curve as the quantitation method. The standard curve method is
used to determine the absolute target quantity in samples. When setting up your
reaction plate, the standard curve method requires targets, standards, and samples.
• Select the reagents you want to use:
–Select TaqMa n
®
Reagents if you want to use TaqMan reagents to detect
amplification and quantify the amount of target in the samples. TaqMan reagents
consist of two primers and a TaqMan
®
probe. The primers are designed to
amplify the target. The TaqMan probe is designed to hybridize to the target and
generate fluorescence signal when the target is amplified.
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA
™
dye
as a reporter or quencher with the StepOne
™
system. TAMRA dye may be used as a
reporter or quencher with the StepOnePlus
™
system.
1
2
3
4

Chapter 2 Design the Experiment
Define the Methods and Materials
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
24
Notes
–Select SYBR
®
Green Reagents if you want to use SYBR Green reagents to
detect amplification and quantify the amount of target in the samples. SYBR
Green reagents consist of two primers and SYBR Green dye. The primers are
designed to amplify the target. The SYBR Green dye generates fluorescence
signal when it binds to double-stranded DNA. SYBR Green dye is often part of
the SYBR Green master mix added to the reaction. If you use SYBR Green dye:
Select the Include Melt Curve checkbox to perform melt curve analysis of the
amplified target.
Select Standard as the ramp speed. A Fast master mix for SYBR Green reagents
is not available from Applied Biosystems.
Note: You can use other fluorescent-based reagents on the StepOne and
StepOnePlus systems, however, you must design your experiment using Advanced
Setup instead of the Design Wizard.
• Select the appropriate ramp speed for the instrument run:
–Select Fast (~ 40 minutes to complete a run) if you are using Fast reagents for
the PCR reactions.
–Select Standard (~ 2 hours to complete a run) if you are using standard
reagents for the PCR reactions.
• Select the appropriate PCR template:
–Select cDNA (complementary DNA) if you are performing 2-step RT-PCR, and
you have already performed reverse transcription to convert the RNA to cDNA.
You are adding complementary DNA to the PCR reactions.
–Select RNA if you are performing 1-step RT-PCR. You are adding total RNA or
mRNA to the PCR reactions.
Note: To use the Fast ramp speed with RNA templates, you must design your
experiment using Advanced Setup instead of the Design Wizard.
–Select gDNA (genomic DNA) if you have already extracted the gDNA from
tissue or sample. You are adding purified genomic DNA to the PCR reactions.
For More
Information
For more information on:
• Completing the Methods & Materials screen, access the StepOne Software Help by
clicking or pressing F1.
• Using Advanced Setup, see “Advanced Setup Workflow” on page 116.
• Using other quantitation methods, refer to the Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems Getting Started Guide for Relative Standard
Curve and Comparative C
T
Experiments.
• TaqMan and SYBR Green reagents, refer to the Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems Reagent Guide.
• PCR, including singleplex vs. multiplex PCR and 1-step vs. 2-step RT PCR, refer to
the Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time PCR Systems
Reagent Guide.

Chapter 2 Design the Experiment
Set Up the Targets
25
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Set Up the Targets
On the Targets screen, enter the number of targets you want to quantify in the PCR
reaction plate, then set up the assays for each target.
About the
Example
Experiment
In the standard curve example experiment:
•One target is quantified in the reaction plate.
• The Set Up Standards checkbox is selected. When this checkbox is selected, the
software automatically displays the Standards screen after you complete the Targets
screen. In the Standards screen, you can set up a standard curve for the target assay
(see “Set Up the Standards” on page 27).
• The Target 1 assay is set up for the target you are studying. For the example
experiment, this is the RNase P gene.
Complete the
Targe ts Sc reen
1. Click the How many targets do you want to quantify in the reaction plate? field,
then enter 1.
Note: The number of rows in the target assays table updates with the number you
entered.
2. Select the Set Up Standards checkbox to set up standards for the target assay.
Note: The Set Up Standards checkbox is selected by default.
3. Set up the Target 1 assay:
a. Click the Enter Target Name cell, then enter RNase P.
b. From the Reporter dropdown menu, select FAM (default).
c. From the Quencher dropdown menu, select NFQ-MGB (default).
d. Leave the default in the Color field.
4. Click Next >.
Note: Leave the (Optional) Enter Gene Name field blank. You can search for the
gene/assay ID when you order your materials (see “Order Materials for the Experiment”
on page 38).

Chapter 2 Design the Experiment
Set Up the Targets
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
26
Notes
Design
Guidelines
When you design your own standard curve experiment:
• Select the Set Up Standards checkbox. Applied Biosystems recommends that you
set up a standard curve for each target assay in the reaction plate.
• Identify each target assay with a unique name and color. You can enter up to 100
characters in the Target Name field.
• Select the reporter dye used in the target assay. In the Methods & Materials screen
on page 22, if you selected:
–TaqMan
®
Reagents, select the dye attached to the 5′ end of the TaqMan probe.
– SYBR
®
Green Reagents, select SYBR.
• Select the quencher used in the target assay. In the Methods & Materials screen on
page 22, if you selected:
–TaqMan
®
Reagents, select the quencher attached to the 3′ end of the TaqMan
probe.
– SYBR
®
Green Reagents, select None.
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA dye as
a reporter or quencher with the StepOne system. TAMRA dye may be used as a
reporter or quencher with the StepOnePlus system.
For More
Information
For more information on completing the Targets screen, access the StepOne Software
Help by clicking or pressing F1.
2
3a
1
3b 3c 3d

Chapter 2 Design the Experiment
Set Up the Standards
27
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Set Up the Standards
On the Standards screen, enter the number of points and replicates for all standard curves
in the reaction plate. For each standard curve, enter the starting quantity and select the
serial factor.
About the
Example
Experiment
In the standard curve example experiment:
• One standard curve is set up for the target (RNase P). The standard used for the
standard dilution series contains known quantities of the RNase P gene. Because a
single gene is being studied, only one standard curve is required.
• Five points are used in the standard curve.
• Three replicates are used for each point. Replicates are identical reactions,
containing identical reaction components and volumes.
• The starting quantity is 10,000 copies and the serial factor is 1:2.
Complete the
Standards Screen
1. Click the How many points do you need for each standard curve? field, then
enter 5.
2. Click the How many replicates do you need for each point? field, then enter 3.
3. Define the range of standard quantities for the RNase P assay:
a. Click the Enter Starting Quantity field, then enter 10000.
b. From the Select Serial Factor dropdown menu, select 1:2.
4. Review the Standard Curve Preview pane. The standard curve has the following
points: 10000, 5000, 2500, 1250, and 625.
5. Click Next >.
1
2
3a
3b
4

Chapter 2 Design the Experiment
Set Up the Standards
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
28
Notes
Design
Guidelines
When you design your own standard curve experiment:
• Set up a standard curve for each target in the reaction plate. The targets are
previously defined on the Targets screen (“Set Up the Targets” on page 25).
• Enter the number of points for each standard curve in the reaction plate. Applied
Biosystems recommends at least five dilution points for each standard curve.
• Enter the number of replicates (identical reactions) for each point in the standard
curve. Applied Biosystems recommends three replicates for each point.
• Because the range of standard quantities affects the amplification efficiency
calculations, carefully consider the appropriate range of standard quantities for your
assay:
– For more accurate measurements of amplification efficiency, use a broad range
of standard quantities, spanning between 5 and 6 logs. If you specify a broad
range of quantities for the standards, you need to use a PCR product or a highly
concentrated template, such as a cDNA clone.
– If you have a limited amount of cDNA template and/or if the target is a low-copy
number transcript, or known to fall within a given range, a narrow range of
standard quantities may be necessary.
• The serial factor is used to calculate the quantities in all points of the standard curve.
If your starting quantity is the highest quantity, select a dilution factor such as 1:2,
1:3, and so on. If your starting quantity is the lowest quantity, select a concentration
factor such as 2✕, 3✕, and so on.
For More
Information
For more information on:
• Completing the Standards screen, access the StepOne Software Help by clicking
or pressing F1.
• Amplification efficiency, refer to the Amplification Efficiency of TaqMan
®
Gene
Expression Assays Application Note.

Chapter 2 Design the Experiment
Set Up the Samples
29
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Set Up the Samples
On the Samples screen, enter the number of samples, replicates, and negative controls to
include in the reaction plate, enter the sample names, then select the sample/target
reactions to set up.
About the
Example
Experiment
In the standard curve example experiment:
• Two samples are used: genomic DNA from two populations. The samples contain
unknown quantities of the target (RNase P).
• Three replicates are used. The replicates are identical reactions, containing identical
reaction components and volumes.
• Three negative controls are used. The negative control reactions contain water
instead of sample and should not amplify.
Complete the
Samples Screen
1. Click the How many samples do you want to test in the reaction plate? field,
then enter 2.
Note: The number of rows in the samples table updates with the number you
entered.
2. Click the How many replicates do you need? field, then enter 3.
3. Click the How many negative controls do you need for each target assay? field,
then enter 3.
4. Set up Sample 1:
a. Click the Enter Sample Name field, then enter pop1 (for population 1).
b. Leave the default in the Color field.
5. Set up Sample 2:
a. Click the Enter Sample Name field, then enter pop2 (for population 2).
b. Leave the default in the Color field.
6. Select All Sample/Target Reactions to test all targets in all samples.
7. In the Well Count pane, confirm there are:
• 6 Unknown wells
• 15 Standard wells
• 3 Negative control wells
• 24 Empty wells
8. In the View Plate Layout tab:
a. From the Arrange Plate by dropdown menu, select Rows (default).

Chapter 2 Design the Experiment
Set Up the Samples
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
30
Notes
b. From the Place Negative Controls in dropdown menu, select Upper Left
(default).
9. Click Next >.
Design
Guidelines
When you design your own standard curve experiment:
• Identify each sample with a unique name and color. You can enter up to 100
characters in the Sample Name field.
• Enter the number of replicates (identical reactions) to set up. Applied Biosystems
recommends three replicates for each sample reaction.
• Enter the number of negative control reactions to set up. Applied Biosystems
recommends three negative control reactions for each target assay.
• Select which targets to test in the samples:
–Select All Sample/Target Reactions to test all targets in all samples.
–Select Specify Sample/Target Reactions to specify the targets to test in each
sample.
Note: When you use the Design Wizard to set up a standard curve experiment, you
can set up only singleplex reactions (amplification and detection of one target per
well). If you want to set up a standard curve experiment with multiplex reactions
(amplification and detection of two or more targets per well), design your
experiment using Advanced Setup instead of the Design Wizard.
• If you are running the experiment on a StepOnePlus instrument and plan to edit the
Run Method (page 31) to set a different temperature for one or more of the VeriFlex
blocks, you need to:
a. Design your experiment using Advanced Setup instead of the Design Wizard.
1
2
3
4
5
6
7
8a
8b

Chapter 2 Design the Experiment
Set Up the Run Method
31
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
b. In the Plate Setup screen, select the Assign Targets and Samples tab, select
the View Plate Layout tab, then select the Enable VeriFlex
™
Block checkbox.
IMPORTANT! If you do not select the Enable VeriFlex
™
Block checkbox in the
Plate Setup screen, you will not be able to set a different temperature for one or
more of the VeriFlex blocks in the Run Method screen (page 31).
For More
Information
For more information on:
• Completing the Samples screen, access the StepOne Software Help by clicking
or pressing F1.
• Using Advanced Setup, see “Advanced Setup Workflow” on page 116.
Set Up the Run Method
On the Run Method screen, review the reaction volume and the thermal profile for the
default run method. If needed, you can edit the default run method or replace it with one
from the Run Method library.
About the
Example
Experiment
In the standard curve example experiment, the default run method is used with one
change: The reaction volume per well is changed from 20 μL to 25 μL.
Review the Run
Method Screen
1. Click either the Graphical View tab (default) or Tabu la r View tab.
2. Click the Reaction Volume Per Well field, then enter 25 μL.
3. Make sure the thermal profile displays the holding and cycling stages shown below.
4. Click Next >.

Chapter 2 Design the Experiment
Set Up the Run Method
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
32
Notes
Design
Guidelines
When you design your own standard curve experiment:
• Enter a number from 10 to 30 for the reaction volume/well. The StepOne and
StepOnePlus systems support reaction volumes from 10 to 30 µL.
• Review the thermal profile:
– Make sure the thermal profile is appropriate for your reagents.
– If you are performing 1-step RT-PCR, include a reverse transcription step.
If your experiment requires a different thermal profile, edit the thermal profile or
replace the run method with one from the Run Method library. The Run Method
library is included in the StepOne software.
• If you are running the experiment on a StepOnePlus instrument and you want to set
a different temperature for one or more of the VeriFlex blocks, you need to:
a. Design your experiment using Advanced Setup instead of the Design Wizard.
b. In the Plate Setup screen (page 30), select the Assign Targets and Samples
tab, select the View Plate Layout tab, then select the Enable VeriFlex
™
Block
checkbox.
IMPORTANT! If you do not select the Enable VeriFlex
™
Block checkbox in
the Plate Setup screen, you will not be able to set a different temperature for
one or more of the VeriFlex blocks in the Run Method screen.
c. In the Run Method screen, select the Graphical View tab.
2
1
3

Chapter 2 Design the Experiment
Review the Reaction Setup
33
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
d. For each VeriFlex
™
block you want to change, click the temperature, then enter
the desired value.
Note: You can set a different temperature for one or more of the VeriFlex blocks, or
set each of the VeriFlex blocks to the same temperature. If neighboring VeriFlex
blocks are not set to the same temperature, the temperature difference must be
between 0.1 and 5.0 °C. The maximum temperature is 99.9 °C.
For More
Information
For more information on:
• The Run Method library or on completing the Run Method screen, access the
StepOne Software Help by clicking or pressing F1.
• Setting temperatures for the VeriFlex blocks, access the StepOne Software Help by
clicking or pressing FI.
• Using Advanced Setup, see “Advanced Setup Workflow” on page 116.
Review the Reaction Setup
On the Reaction Setup screen, select the assay type (if using TaqMan reagents), then
review the calculated volumes for preparing the PCR reactions, standard dilution series,
and sample dilutions. If needed, you can edit the reaction volume, excess reaction
volume, component concentrations, standard concentration, and/or diluted sample
concentration.
IMPORTANT! Perform these steps for each target assay in the reaction plate.
About the
Example
Experiment
In the standard curve example experiment:
• Applied Biosystems RNase P assay is used.
• The reaction volume per well is 25 μL.
• The excess reaction volume is 10%.
• The reaction components are:
–TaqMan
®
Universal PCR Master Mix (2✕) or TaqMan
®
2✕ Universal PCR
Master Mix, No AmpErase
®
UNG
– RNase P Assay Mix (20✕)
– Sample or standard
– Water
• The standard concentration in stock is 20,000 copies/μL.
• The diluted sample concentration is 6.6 ng/μL.
• The sample stock concentration is 100 ng/μL.

Chapter 2 Design the Experiment
Review the Reaction Setup
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
34
Notes
Complete the
Reaction Setup
Screen
Complete the Reaction Mix Calculations Tab
1. Select the Reaction Mix Calculations tab (default).
2. From the Select Target pane, select RNase P (default).
3. From the Assay Type dropdown menu, select Inventoried/Made to Order.
4. Make sure the Reaction Volume Per Well field displays 25 µL.
5. Make sure the Excess Reaction Volume field displays 10%.
6. In the Reactions for RNase P pane:
a. Make sure the Master Mix Concentration field displays 2.0✕.
b. Make sure the Assay Mix Concentration field displays 20.0✕.
c. Review the components and calculated volumes for the PCR reactions:
Component Volume (µL) for 1 Reaction
Master Mix (2.0✕)12.5
Assay Mix (20.0✕)1.3
Sample (10✕) or Standard 2.5
‡
‡ The sample or standard volume is limited to 10% of the total reaction
volume.
H
2
O8.8
Total Volume 25.0
1
2
3 54
6b
6a
6c

Chapter 2 Design the Experiment
Review the Reaction Setup
35
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
7. In the Standard Dilution Series for RNase P pane:
a. Click the Standard Concentration in Stock field, then enter 20000.
b. Click the units field, then enter copies per μL.
c. Review the calculated volumes for preparing the standard dilution series:
Complete the Sample Dilution Calculations Tab
1. Select the Sample Dilution Calculations tab.
2. Click the Diluted Sample Concentration (10✕ for Reaction Mix) field, then
enter 6.6.
3. From the unit dropdown menu, select ng/μL (default).
Dilution
Point
Source
Source
Volume
(µL)
Diluent
Volume
(µL)
Total
Volume
(µL)
Standard
Concentration
(copies/µL)
1 (10,000) Stock 3.6 14.5 18.2 4000.0
2 (5,000) Dilution 1 9.1 9.1 18.2 2000.0
3 (2,500) Dilution 2 9.1 9.1 18.2 1000.0
4 (1,250) Dilution 3 9.1 9.1 18.2 500.0
5 (625) Dilution 4 9.1 9.1 18.2 250.0
7b
7a
7c

Chapter 2 Design the Experiment
Review the Reaction Setup
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
36
Notes
4. Review the calculated volumes for the sample dilutions:
Print Reaction Setup Instructions
Print detailed reaction setup instructions, then save the instructions for Chapter 3,
“Prepare the Reactions.”
1. Click Print Reaction Setup.
2. In the dialog box, select:
• Detailed Reaction Setup Instructions
• Include Plate Layout
• Use sample color
3. Click Print to send the reaction setup instructions to your printer.
Sample Name
Stock
Concentration
(ng/µL)
Sample
Volume (µL)
Diluent
Volume (µL)
Total Volume
of Diluted
Sample (µL)
pop1 100.0 1.0 14.2 15.2
pop2 100.0 1.0 14.2 15.2
1
2
3
4
1

Chapter 2 Design the Experiment
Review the Reaction Setup
37
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
4. Click Next >.
Design
Guidelines
When you design your own standard curve experiment:
• If you are using TaqMan reagents, select the type of assay you are using:
–Select Inventoried/Made to Order if you are using Applied Biosystems
TaqMan
®
Gene Expression Assays (Inventoried or Made to Order) or Applied
Biosystems Custom TaqMan
®
Gene Expression Assays.
–Select Custom if you are designing your own assays with Primer Express
®
software.
• Enter a number from 10 to 30 for the reaction volume/well. The StepOne and
StepOnePlus systems support reaction volumes from 10 to 30 µL.
• Include excess reaction volume to account for the loss that occurs during pipetting.
Applied Biosystems recommends an excess reaction volume of at least 10%.
• Review the reaction mix concentrations for each target: If needed:
– For TaqMan reagents, edit the master mix and assay mix concentrations.
– For SYBR Green reagents, edit the master mix, forward primer, and reverse
primer concentrations.
– For 1-step RT-PCR, edit the reverse transcriptase concentration.
• Review the reaction mix components for each target:
– If you are running Fast PCR reactions, make sure you use Fast master mix in the
PCR reactions.
– If you are running standard PCR reactions, make sure you use standard master
mix in the PCR reactions.
– For 1-step RT-PCR, make sure you include reverse transcriptase in the PCR
reactions and use a specific buffer.
• Review the standard dilution series calculations for each target. If needed, edit the
standard concentration in stock (including units).
Note: For the Standard Concentration in Stock units field, you can select ng or μg
from the dropdown menu or you can enter another unit in the field (for example,
copies, IU, [International Units], nmol, pg, and so on). The table updates according
to your entry.
2
3
Chapter 2 Design the Experiment
Order Materials for the Experiment
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
38
Notes
• Review the sample dilution calculations for each sample. If needed, edit the diluted
sample concentration (including units) and stock concentration.
For More
Information
For more information on:
• Completing the Reaction Setup screen, access the StepOne Software Help by
clicking or pressing F1.
• Applied Biosystems assays, refer to the:
– TaqMan
®
Gene Expression Assays Protocol
– Custom TaqMan
®
Gene Expression Assays Protocol
Order Materials for the Experiment
On the Materials List screen, review the list of materials recommended to prepare the
PCR reaction plate. (Optional) Print the materials list, create a shopping list, then order
the recommended materials from the Applied Biosystems Store.
Note: To access the Applied Biosystems Store, you need to have an Internet connection.
Product availability and pricing may vary according to your region or country. Online
ordering through the Applied Biosystems Store is not available in all countries. Contact
your local Applied Biosystems representative for help.
Note: The StepOne software recommends the materials to order based on your
experiment design. It is assumed that you will design your experiment, order your
materials, then prepare (Chapter 3) and run (Chapter 4) the reaction plate when your
materials arrive.
About the
Example
Experiment
In the standard curve example experiment, the recommended materials are:
• MicroAmp
™
Fast Optical 48-Well Reaction Plate
• MicroAmp
™
48-Well Optical Adhesive Film
• MicroAmp
™
96-Well Support Base
•TaqMan
®
Universal PCR Master Mix (2✕) or TaqMan
®
2✕ Universal PCR Master
Mix, No AmpErase
®
UNG

Chapter 2 Design the Experiment
Order Materials for the Experiment
39
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
• Applied Biosystems RNase P assay
Note: The human RNase P FAM
™
dye-labeled MGB probe is not available as a
TaqMan
®
Gene Expression Assay. It can be ordered as a Custom TaqMan
®
Gene
Expression Assay (PN 4331348).
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA dye as
a reporter or quencher with the StepOne system. TAMRA dye may be used as a
reporter or quencher with the StepOnePlus system.
Note: The example experiment was created for a StepOne instrument. If you selected the
StepOnePlus instrument in the Experiment Properties screen (page 20), the 96-well
consumables (for example, the MicroAmp
™
Fast Optical 96-Well Reaction Plate) are
listed in place of the 48-well consumables.
Complete the
Materials List
Screen
1. For the example experiment, leave the Find Assay pane empty.
The human RNase P FAM dye-labeled MGB probe is not available as a TaqMan
Gene Expression Assay and cannot be ordered from the Order Materials List screen.
If desired, you can order it as a Custom TaqMan Gene Expression Assay
(PN 4331348). Refer to the Custom TaqMan
®
Gene Expression Assays Protocol.
Note: When you design your own standard curve experiment, see “Design
Guidelines” on page 41 for information on how to complete the Find Assay pane.
2. From the Display dropdown menu, select All Items (default), then review the
recommended materials. If needed, use the scroll bar at right to see all items.
Note: For more information on a specific item, click the part number link. You will
be connected to the product information page on Applied Biosystems Store. To
access the Applied Biosystems Store, you need to have an Internet connection.
3. (Optional) Click Print Materials List to send the materials list to your printer.
4. (Optional) Create a shopping list:
a. Select the checkbox next to each of the following items:
• MicroAmp
™
Fast Optical 48-Well Reaction Plate
• MicroAmp
™
48-Well Optical Adhesive Film
• MicroAmp
™
96-Well Support Base
•TaqMan
®
Universal PCR Master Mix (2✕) or TaqMan
®
2✕ Universal
PCR Master Mix, No AmpErase
®
UNG
b. Click Add Selected Items to Shopping List.

Chapter 2 Design the Experiment
Order Materials for the Experiment
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
40
Notes
5. (Optional) Create a shopping basket on the Applied Biosystems Store:
Note: To access the Applied Biosystems Store, you need to have an Internet
connection. Product availability and pricing may vary according to your region or
country. Online ordering through the Applied Biosystems Store is not available in all
countries. Contact your local Applied Biosystems representative for help.
a. Check that the Experiment Shopping List contains the desired materials and
that the quantities are correct, then click Order Materials in List.
b. In the Order Materials - Log In dialog box, enter your user name and password
for the Applied Biosystems Store, then click Login and Submit.
Note: If you do not have an account with the Applied Biosystems Store, click
Register Now to create an account.
4b
2
4a
5a
3
5b

Chapter 2 Design the Experiment
Order Materials for the Experiment
41
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
c. When you are connected to the Applied Biosystems Store, follow the prompts
to complete your order.
6. Go to “Finish the Design Wizard” on page 42.
Design
Guidelines
When you design your own standard curve experiment:
• Select all the materials you require for your experiment and add them to your
shopping list.
• To access the Applied Biosystems Store:
– Confirm that your computer has an Internet connection.
– Applied Biosystems recommends the following browsers and Adobe
®
Acrobat
®
Reader versions to use the Applied Biosystems web site:
Note: Make sure that cookies and Java Script are turned on for the web site to
function correctly.
• To find your assay on the Applied Biosystems Store, complete the Find Assay pane:
a. Click the Enter Gene Name field, enter the gene name, then click Find Assay.
b. In the Find Assay Results dialog box, select your assay.
c. Click Apply Assay Selection.
For More
Information
For more information on completing the Materials List screen, access the StepOne
Software Help by clicking or pressing F1.
Desktop
Operating
System
Netscape
®
Navigator
Microsoft
®
Internet
Explorer
Macintosh
®
Safari
Adobe
®
Acrobat
®
Reader
Windows
®
2000/NT/XP/Vista
v6.x or later v6.x or later Not applicable v4.0 or later
Macintosh
®
OS
9+ or later
Not supported Not supported v2.0.4 or later v4.0 or later

Chapter 2 Design the Experiment
Finish the Design Wizard
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
42
Notes
Finish the Design Wizard
To finish the Design Wizard, review the plate layout, then select an exit option.
About the
Example
Experiment
The StepOne software automatically selects locations for the wells in the reaction plate.
In the standard curve example experiment:
• The wells are arranged as shown below.
Note: The example experiment was created for a StepOne instrument. If you
selected the StepOnePlus instrument in the Experiment Properties screen (page 20),
your reaction plate layout will differ from the layout shown above. The software
displays a 96-well reaction plate layout for the StepOnePlus instrument. For an
example of the 96-well reaction plate layout, see page 11.
• The experiment is saved as is and closed.
Note: For the example experiment, do not perform the run at this time.
Finish the
Design Wizard
1. At the bottom of the StepOne software screen, click Finish Designing Experiment.
2. In the Review Plate for Experiment window, review the plate layout. Make sure
there are:
• 6 Unknown wells
• 15 Standard wells
• 3 Negative control wells
• 24 Empty wells
Note: If the plate layout is incorrect, click Return to the Wizard and check your
entered values.

Chapter 2 Design the Experiment
Finish the Design Wizard
43
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
3. Click Save Experiment.
4. In the Save Experiment dialog box, click Save to accept the default file name and
location. The example experiment is saved and closed, and you are returned to the
Home screen.
Note: By default, the example experiment is saved to the <drive>:\Applied
Biosystems\<software name>\experiments folder.
2
3
4

Chapter 2 Design the Experiment
Finish the Design Wizard
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
44
Notes
Design
Guidelines
When you design your own standard curve experiment:
• In the Review Plate for Experiment window, select the appropriate exit option:
• By default, experiments are saved to the <drive>:\Applied Biosystems\<software
name>\experiments folder. To change the:
– Save location for a specific experiment, navigate to the desired location using the
Save Experiment dialog box.
– Default save location, select ToolsPreferences, then select the General tab
(default). In the Default Data Folder field, browse to the desired location.
For More
Information
For more information on using Advanced Setup, see “Advanced Setup Workflow” on
page 116.
Click If you want to...
Save Experiment save and close the experiment without making any
further changes or starting the run.
Start Run for This Experiment save the experiment and start the run. Make sure the
reaction plate is loaded in the instrument.
Edit Plate Layout • use advanced setup to edit the plate layout.
•(StepOnePlus instrument only) set a different
temperature for one or more of the VeriFlex blocks
using Advanced Setup.
Create Another Experiment Using
the Design Wizard
save and close the current experiment, then create
another experiment using the Design Wizard.
Return to the Wizard return to the experiment to make changes using the
Design Wizard.
Chapter 3
45
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Prepare the Reactions
This chapter covers:
■ Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 46
■ Prepare the Sample Dilutions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 47
■ Prepare the Standard Dilution Series . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 49
■ Prepare the Reaction Mix . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 51
■ Prepare the Reaction Plate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 53
Note: For more information about any of the topics discussed in this guide, access the
Help from within Applied Biosystems StepOne
™
Real-Time PCR Software v2.0 by
pressing F1, clicking in the toolbar, or selecting HelpStepOne Software Help.

Chapter 3 Prepare the Reactions
Chapter Overview
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
46
Notes
Chapter Overview
This chapter explains how to prepare the PCR reactions for the standard curve example
experiment and provides guidelines for how to prepare the PCR reactions for your own
standard curve experiment.
Example
Experiment
Workflow
The workflow for preparing the PCR reactions for the example experiment provided with
this getting started guide is shown below.
Design the Experiment (Chapter 2)
Start Experiment
Run the Experiment (Chapter 4)
Analyze the Experiment (Chapter 5)
End Experiment
Prepare the Reactions (Chapter 3)
1. Prepare the sample dilutions.
2. Prepare the standard dilution series.
3. Prepare the reaction mix for each target assay.
4. Prepare the reaction plate.

Chapter 3 Prepare the Reactions
Prepare the Sample Dilutions
47
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Prepare the Sample Dilutions
Perform sample dilutions before adding the samples to the final reaction mix. Dilute the
samples using the volumes that were calculated by the StepOne
™
software (“Complete
the Sample Dilution Calculations Tab” on page 35).
About the
Example
Experiment
For the standard curve example experiment:
• Sample dilutions are necessary because the sample volume is limited to 10% of the
total reaction volume in the StepOne software. The total reaction volume is
25 μL/reaction, so the sample volume is 2.5 μL/reaction.
• The stock concentration is 100 ng/μL. After diluting the sample according to the
Sample Dilutions Calculations table, the sample will have a concentration of
6.6 ng/μL. This is a 10✕ concentration when adding 2.5 μL to the final reaction mix
volume of 25 μL. You will have a 1✕ concentration in the final reaction.
• The volumes calculated in the software are:
Required
Materials
• Water to dilute the sample
• Microcentrifuge tubes
• Pipettors
• Pipette tips
• Sample stock
•Vortexer
• Centrifuge
Prepare the
Sample Dilutions
1. Label a separate microcentrifuge tube for each diluted sample:
• Population 1
• Population 2
2. Add the required volume of water (diluent) to each empty tube:
Sample Name
Stock
Concentration
(ng/µL)
Sample
Volume (µL)
Diluent
Volume (µL)
Total Volume
of Diluted
Sample (µL)
pop1 100.0 1.0 14.2 15.2
pop2 100.0 1.0 14.2 15.2
Tube Samp l e Name
Diluent Volume
(µL)
1 Population 1 14.2
2 Population 2 14.2

Chapter 3 Prepare the Reactions
Prepare the Sample Dilutions
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
48
Notes
3. Add the required volume of sample stock to each tube:
4. Vortex each diluted sample for 3 to 5 seconds, then centrifuge the tubes briefly.
5. Place the diluted samples on ice until you prepare the reaction plate.
Preparation
Guidelines
When you prepare your own standard curve experiment:
• Sample dilutions may be necessary because the sample volume is limited to 10% of
the total reaction volume in the StepOne software. You must perform the sample
dilutions before adding the samples to the final reaction mix.
• For optimal performance of TaqMan
®
Gene Expression Assays or Custom
TaqMan
®
Gene Expression Assays, use 10 to 100 ng of cDNA template per
20-µL reaction. For Fast reagents, Applied Biosystems recommends 10 ng.
• Use TE buffer or water to dilute the sample.
For More
Information
For more information on Applied Biosystems assays, refer to the:
• TaqMan
®
Gene Expression Assays Protocol
• Custom TaqMan
®
Gene Expression Assays Protocol
Tube Samp l e Name
Sample Volume
(µL)
1Population 1 1.0
2Population 2 1.0

Chapter 3 Prepare the Reactions
Prepare the Standard Dilution Series
49
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Prepare the Standard Dilution Series
Prepare the standard dilution series using the volumes that were calculated by the
StepOne software (“Complete the Reaction Mix Calculations Tab” on page 34).
About the
Example
Experiment
For the standard curve example experiment:
• The standard concentration in stock is 20,000 copies/μL.
• The volumes calculated in the software are:
Required
Materials
• Water to dilute the standards
• Microcentrifuge tubes
• Pipettors
• Pipette tips
• Standard stock
•Vortexer
• Centrifuge
Prepare the
Standard Dilution
Series for the
RNase P Assay
1. Label a separate microcentrifuge tube for each standard:
• RNase P Std. 1
• RNase P Std. 2
• RNase P Std. 3
• RNase P Std. 4
• RNase P Std. 5
2. Add the required volume of water (diluent) to each empty tube:
Dilution
Point
Source
Source
Volume
(µL)
Diluent
Volume
(µL)
Tot al
Volume
(µL)
Standard
Concentration
(copies/µL)
1 (10,000) Stock 3.6 14.5 18.2 4000.0
2 (5,000) Dilution 1 9.1 9.1 18.2 2000.0
3 (2,500) Dilution 2 9.1 9.1 18.2 1000.0
4 (1,250) Dilution 3 9.1 9.1 18.2 500.0
5 (625) Dilution 4 9.1 9.1 18.2 250.0
Tube Standard Name
Volume of
Diluent to
Add (µL)
1RNase P Std. 1 14.5
2 RNase P Std. 2 9.1
3 RNase P Std. 3 9.1

Chapter 3 Prepare the Reactions
Prepare the Standard Dilution Series
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
50
Notes
3. Prepare dilution 1 in the RNase P Std. 1 tube:
a. Vortex the stock for 3 to 5 seconds, then centrifuge the tube briefly.
b. Using a new pipette tip, add 3.6 µL of stock to the RNase P Std. 1 tube.
c. Vortex Std. 1 for 3 to 5 seconds, then centrifuge the tube briefly.
4. Prepare dilution 2 in the RNase P Std. 2 tube:
a. Using a new pipette tip, add 9.1 µL of dilution 1 to the RNase P Std. 2 tube.
b. Vortex Std. 2 for 3 to 5 seconds, then centrifuge the tube briefly.
5. Prepare dilution 3 in the RNase P Std. 3 tube:
a. Using a new pipette tip, add 9.1 µL of dilution 2 to the RNase P Std. 3 tube.
b. Vortex Std. 3 for 3 to 5 seconds, then centrifuge the tube briefly.
6. Prepare dilution 4 in the RNase P Std. 4 tube:
a. Using a new pipette tip, add 9.1 µL of dilution 3 to the RNase P Std. 4 tube.
b. Vortex Std. 4 for 3 to 5 seconds, then centrifuge the tube briefly.
7. Prepare dilution 5 in the RNase P Std. 5 tube:
a. Using a new pipette tip, add 9.1 µL of dilution 4 to the RNase P Std. 5 tube.
b. Vortex Std. 5 for 3 to 5 seconds, then centrifuge the tube briefly.
8. Place the standards on ice until you prepare the reaction plate.
Preparation
Guidelines
When you prepare your own standard curve experiment:
• Standards are critical for accurate analysis of run data.
• Any mistakes or inaccuracies in making the dilutions directly affect the quality of
results.
• The quality of pipettors and tips and the care used in measuring and mixing
dilutions affect accuracy.
• Use TE buffer or water to dilute the standards.
4 RNase P Std. 4 9.1
5 RNase P Std. 5 9.1
Tube Standard Name
Volume of
Diluent to
Add (µL)

Chapter 3 Prepare the Reactions
Prepare the Reaction Mix
51
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Prepare the Reaction Mix
Prepare the reaction mix using the components and volumes that were calculated by the
StepOne software (“Complete the Reaction Mix Calculations Tab” on page 34).
Note: The software calculates the volumes for all components for the PCR reactions.
However, when you prepare the reaction mix in this section, include only the master mix,
assay mix, and water. Add the sample or standard when you prepare the reaction plate
(see “Prepare the Reaction Plate” on page 53).
About the
Example
Experiment
For the standard curve example experiment:
• The reaction mix components are:
–TaqMan
®
Universal PCR Master Mix (2✕) or TaqMan
®
2✕ Universal PCR
Master Mix, No AmpErase
®
UNG
– RNase P Assay Mix (20✕)
– Water
• The volumes calculated in the software are:
Note: The sample or standard is not added at this time.
Required
Materials
• Microcentrifuge tubes
• Pipettors
• Pipette tips
• Reaction mix components (listed above)
• Centrifuge
Component Volume (µL) for 1 Reaction
Master Mix (2.0✕)12.5
Assay Mix (20.0✕)1.3
H
2
O8.8
Total Volume 22.6

Chapter 3 Prepare the Reactions
Prepare the Reaction Mix
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
52
Notes
Prepare the
Reaction Mix
CHEMICAL HAZARD. TaqM an
®
Universal PCR Master Mix
(2✕) may cause eye and skin irritation. Exposure may cause discomfort if swallowed or
inhaled. Read the MSDS, and follow the handling instructions. Wear appropriate
protective eyewear, clothing, and gloves.
CHEMICAL HAZARD. TaqM an
®
2✕ Universal PCR Master
Mix, No AmpErase
®
UNG may cause eye and skin irritation. Exposure may cause
discomfort if swallowed or inhaled. Read the MSDS, and follow the handling
instructions. Wear appropriate protective eyewear, clothing, and gloves.
1. Label an appropriately sized tube for the reaction mix: RNase P Reaction Mix.
2. For the RNase P assay, add the required volumes of each component to the tube:
3. Mix the reaction mix by gently pipetting up and down, then cap the tube.
4. Centrifuge the tube briefly to remove air bubbles.
5. Place the reaction mix on ice until you prepare the reaction plate.
Preparation
Guidelines
When you prepare your own standard curve experiment:
• If your experiment includes more than one target assay, prepare the reaction mix for
each target assay separately.
• Include excess volume in your calculations to provide excess volume for the loss
that occurs during reagent transfers. Applied Biosystems recommends an excess
volume of at least 10%.
• Include all required components.
• Prepare the reagents according to the manufacturer’s instructions.
• Keep the assay mix protected from light, in the freezer, until you are ready to use it.
Excessive exposure to light may affect the fluorescent probes.
• Prior to use:
– Mix the master mix thoroughly by swirling the bottle.
– Resuspend the assay mix by vortexing, then centrifuge the tube briefly.
Component
Volume (µL) for
1 Reaction
Volume (µL) for
24 Reactions
(Plus 10% Excess)
Ta qM an
®
Universal PCR Master
Mix (2✕) or TaqMan
®
2✕
Universal PCR Master Mix, No
AmpErase
®
UNG
12.5 330.0
RNase P Assay Mix (20✕) 1.3 34.3
Water 8.8 232.3
Total Reaction Mix Volume 22.6 596.6

Chapter 3 Prepare the Reactions
Prepare the Reaction Plate
53
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
– Thaw any frozen samples by placing them on ice. When thawed, resuspend the
samples by vortexing, then centrifuge the tubes briefly.
For More
Information
For more information on preparing the reaction mix, refer to the protocol appropriate for
the reagents you are using in the PCR reactions:
• TaqMan
®
Gene Expression Assays Protocol
• Custom TaqMan
®
Gene Expression Assays Protocol
Prepare the Reaction Plate
Prepare the reactions for each replicate group, then transfer them to the reaction plate.
Use the plate layout displayed in the StepOne software.
About the
Example
Experiment
For the standard curve example experiment:
•A MicroAmp
™
Fast Optical 48-Well Reaction Plate is used.
• The reaction volume is 25 μL/well.
• The reaction plate contains:
– 6 Unknown wells
– 15 Standard wells
– 3 Negative control wells
– 24 Empty wells
• The plate layout automatically generated by the StepOne software is used:

Chapter 3 Prepare the Reactions
Prepare the Reaction Plate
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
54
Notes
Note: The example experiment was created for a StepOne instrument. If you
selected the StepOnePlus instrument in the Experiment Properties screen (page 20),
your reaction plate layout will differ from the layout shown above. The software
displays a 96-well reaction plate layout for the StepOnePlus instrument. For an
example of the 96-well reaction plate layout, see page 11.
Required
Materials
• Microcentrifuge tubes
• Pipettors
• Pipette tips
• RNase P reaction mix (from page 52)
• Water
• Standards (from page 49)
• Samples (from page 47)
• MicroAmp
™
Fast Optical 48-Well Reaction Plate
• MicroAmp
™
48-Well Optical Adhesive Film
• Centrifuge
Prepare the
Reaction Plate
1. Prepare the negative control reactions for the target:
a. To an appropriately sized tube, add the volumes of reaction mix and water
listed below.
b. Mix the reaction by gently pipetting up and down, then cap the tube.
Tube Reaction Mix
Reaction Mix
Volume (μL)
Water Volume
(μL)
1 RNase P reaction
mix
74.6 8.3

Chapter 3 Prepare the Reactions
Prepare the Reaction Plate
55
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
c. Centrifuge the tube briefly to remove air bubbles.
d. Add 25 µL of the negative control reaction to the appropriate wells in the
reaction plate.
2. For each replicate group, prepare the standard reactions:
a. To appropriately sized tubes, add the volumes of reaction mix and standard
listed below.
b. Mix the reactions by gently pipetting up and down, then cap the tubes.
c. Centrifuge the tubes briefly to remove air bubbles.
d. Add 25 µL of the standard reaction to the appropriate wells in the reaction
plate.
3. For each replicate group, prepare the reactions for the unknowns:
a. To appropriately sized tubes, add the volumes of reaction mix and sample
listed below.
b. Mix the reactions by gently pipetting up and down, then cap the tubes.
c. Centrifuge the tubes briefly to remove air bubbles.
d. Add 25 µL of the unknown (sample) reaction to the appropriate wells in the
reaction plate.
Tube
Standard
Reaction
Reaction
Mix
Reaction
Mix
Volume
(μL)
Standard
Standard
Volume
(μL)
1RNase P
Std 1
RNase P
reaction mix
74.6 RNase P
Std 1
8.3
2RNase P
Std 2
RNase P
reaction mix
74.6 RNase P
Std 2
8.3
3RNase P
Std 3
RNase P
reaction mix
74.6 RNase P
Std 3
8.3
4RNase P
Std 4
RNase P
reaction mix
74.6 RNase P
Std 4
8.3
5RNase P
Std 5
RNase P
reaction mix
74.6 RNase P
Std 5
8.3
Tube
Unknown
Reaction
Reaction
Mix
Reaction
Mix
Volume
(μL)
Sample
Sample
Volume
(μL)
1RNase P
pop1
RNase P
reaction mix
74.6 pop1 8.3
2RNase P
pop2
RNase P
reaction mix
74.6 pop2 8.3

Chapter 3 Prepare the Reactions
Prepare the Reaction Plate
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
56
Notes
4. Seal the reaction plate with optical adhesive film.
5. Centrifuge the reaction plate briefly to remove air bubbles.
6. Confirm that the liquid is at the bottom of each well of the reaction plate. If not,
centrifuge the reaction plate again at a higher speed and for a longer period of time.
IMPORTANT! Do not allow the bottom of the reaction plate to become dirty. Fluids
and other contaminants that adhere to the bottom of the reaction plate can
contaminate the sample block(s) and cause an abnormally high background signal.
7. Until you are ready to perform the run, place the reaction plate on ice in the dark.
Preparation
Guidelines
When you prepare your own standard curve experiment:
• Make sure you use the appropriate consumables.
• Make sure the arrangement of the PCR reactions matches the plate layout displayed
in the StepOne software. You can either:
– Accept the plate layout automatically generated by the software.
or
– Use Advanced Setup to change the plate layout in the software.
• If you use optical adhesive film to seal your reaction plates, seal each reaction plate
as follows:
Correct Incorrect
Liquid is at the
bottom of the well.
• Not centrifuged with enough force, or
• Not centrifuged for enough time
Action
Example
StepOne
™
System StepOnePlus
™
System
1. Place the reaction plate onto the center of the 96-well base. Be sure the reaction plate is flush with the top surface of the
96-well base.
2. Load the reaction plate as desired.
3. Remove a single optical adhesive film (film) from the box.
• For the StepOne system reaction plate, bend both end-tabs
upward. Hold the film backing side up.
• For the StepOnePlus system reaction plate, fold back one of
the end-tabs. Hold the film backing side up.

Chapter 3 Prepare the Reactions
Prepare the Reaction Plate
57
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
For More
Information
For more information on:
• Preparing the reaction plate, refer to the protocol appropriate for the reagents you
are using in the PCR reactions:
–TaqMan
®
Gene Expression Assays Protocol
– Custom TaqMan
®
Gene Expression Assays Protocol
• Consumables, see “Supported Consumables” on page 4.
• Using Advanced Setup to change the plate layout, see “Advanced Setup Workflow”
on page 116.
4. In one swift movement, peel back the white protective backing
from the center sealing surface. Do not touch the center
sealing surface.
IMPORTANT! Improper peeling of the optical adhesive film may
result in haziness, but will not affect results. Haziness will
disappear when the film comes into contact with the heated cover
in the instrument.
5. Holding the film by the end-tabs, lower the film onto the
reaction plate (adhesive side facing the reaction plate). Be sure
the film is completely covering all wells of the reaction plate.
6. While applying firm pressure, move the applicator slowly
across the film, horizontally and vertically, to ensure good
contact between the film and the entire surface of the reaction
plate.
7. While using the applicator to hold the edge of the film in place,
grasp one end of the end-tab and pull up and away sharply.
Repeat for the other end-tab.
8. To ensure a tight, evaporation-free seal:
a. Repeat step 6.
b. While applying firm pressure, run the edge of the applicator along all four sides of the outside border of the film.
Note: Optical adhesive films do not adhere on contact. The films require the application of pressure to ensure a tight,
evaporation-free seal.
9. Inspect the reaction plate to be sure all wells are sealed. You should see an imprint of all wells on the surface of the film.
Action
Example
StepOne
™
System StepOnePlus
™
System

Chapter 3 Prepare the Reactions
Prepare the Reaction Plate
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
58
Notes

Chapter 4
59
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Run the Experiment
This chapter covers:
■ Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 60
■ Prepare for the Run. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 61
■ (Optional) Enable the Notification Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 63
■ Start the Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 65
■ Monitor the Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 69
■ Unload the Instrument and Transfer the Data. . . . . . . . . . . . . . . . . . . . . . . . . . . . . 76
Note: For more information about any of the topics discussed in this guide, access the
Help from within Applied Biosystems StepOne
™
Real-Time PCR Software v2.0 by
pressing F1, clicking in the toolbar, or selecting HelpStepOne Software Help.

Chapter 4 Run the Experiment
Chapter Overview
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
60
Notes
Chapter Overview
This chapter explains how to perform a run on the Applied Biosystems StepOne
™
and
StepOnePlus
™
Real-Time PCR Systems.
Example
Experiment
Workflow
The workflow for running the example experiment provided with this getting started
guide is shown below.
Design the Experiment (Chapter 2)
Start Experiment
Prepare the Experiment (Chapter 3)
Analyze the Experiment (Chapter 5)
End Experiment
Run the Experiment (Chapter 4)
1. Prepare for the run.
2. (Optional) Enable the notification settings.
3. Start the run.
4. Monitor the run.
5. Unload the instrument and transfer the data.

Chapter 4 Run the Experiment
Prepare for the Run
61
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Prepare for the Run
Prepare for the run by opening the example experiment file you created in Chapter 2,
then loading the sealed reaction plate into the StepOne
™
or StepOnePlus
™
instrument.
Open the
Example
Experiment
1. Double-click (StepOne software shortcut) or select
StartAll ProgramsApplied BiosystemsStepOne Software
<software name>
where <software name> is the current version of the StepOne software.
2. From the Home screen, click Open.
3. In the Open dialog box, navigate to the experiments folder (default):
<drive>:\Applied Biosystems\<software name>\experiments
4. Double-click Standard Curve Example to open the example experiment file you
created in Chapter 2.
2
4
3

Chapter 4 Run the Experiment
Prepare for the Run
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
62
Notes
Load the
Reaction Plate
Into the
Instrument
PHYSICAL INJURY HAZARD. During instrument operation, the
temperature of the sample block(s) can exceed 100 °C. If the instrument has been used
recently, keep your hands away until the sample block(s) reaches room temperature.
IMPORTANT! Wear powder-free gloves when you handle the
reaction plate
.
1. Open the instrument drawer.
2. Place the reactions in the sample block(s):
• If using a reaction plate: Place the reaction plate in the sample block(s) with
well A1 at the back-left corner.
• If using reaction tube strips: Place the tray containing the tube strips in the
sample block(s).
• If using reaction tubes: Place the tray containing the tubes in the sample
block(s).
StepOne instrument
sample block
StepOnePlus
instrument VeriFlex
™
Sample Blocks

Chapter 4 Run the Experiment
(Optional) Enable the Notification Settings
63
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
IMPORTANT! For optimal performance with partial loads:
StepOnePlus instruments - Load at least 16 tubes and arrange them in:
• Adjacent columns of 8 tubes, using rows A through H. For example, fill wells
in column 1 (rows A through H) and column 2 (rows A through H).
or
• Adjacent rows of 8 tubes, using columns 3 through 10. For example, fill wells
in row A (columns 3 through 10) and row B (columns 3 through 10).
StepOne instruments - Load at least 4 tubes in the sample block.
3. Close the instrument drawer carefully.
(Optional) Enable the Notification Settings
Enable the notification settings so that the StepOne software alerts you via email when
the StepOne or StepOnePlus instrument begins and completes the run, or if an error
occurs during the run. Enabling the notifications settings feature is optional and does not
affect the performance of the StepOne
™
and StepOnePlus
™
systems or the duration of
the run.
IMPORTANT! The notification settings feature is available only if the computer that you
are using is running the StepOne or StepOnePlus instrument and is connected to an
Ethernet network.
Note: The notification system is also available to computers that are monitoring a
StepOne or StepOnePlus instrument remotely. For more information, see “Remote
Monitor” on page 73.
About the
Example
Experiment
In the example experiment:
• The StepOne software is set up to send notifications to three users (scientist,
supervisor, and technician at mycompany.com) when the StepOne or StepOnePlus
system ends the run and if it encounters any errors during operation.
• The example SMTP server (www.mycompany.com) is set up for Secure Sockets
Layer (SSL) encryption and requires authentication for use.

Chapter 4 Run the Experiment
(Optional) Enable the Notification Settings
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
64
Notes
Set Up the
Notification
Settings
1. In the StepOne software, click Run in the navigation pane.
2. Click Notification Settings.
3. Select Ye s for Enable Notifications.
4. Select the events that will trigger notifications:
a. Select Instrument Error.
b. Select Run Completed.
5. In the Enter e-mail addresses for notifications field, enter:
scientist@mycompany.com, supervisor@mycompany.com,
technician@mycompany.com.
6. In the Outgoing Mail Server (SMTP) field, enter smtp.mycompany.com.
7. Set the authentication settings:
a. Select Ye s for Server requires authentication.
b. In the User Name field, enter Example User.
c. In the Password field, enter password.
Run Guidelines
When you set up the StepOne or StepOnePlus system for automatic notification:
• Your system must be set up for network use. Refer to the Applied Biosystems
StepOne
™
and StepOnePlus
™
Real-Time PCR Systems Installation, Networking,
and Maintenance Guide.
• Select the events for which you want to receive e-mail notifications:
– Instrument Error – When selected, recipients are e-mailed all errors
encountered by the instrument during each run.
– Run Started – When selected, recipients are e-mailed every time the instrument
starts a run.
3
5
6
7a
4
7b
7c

Chapter 4 Run the Experiment
Start the Run
65
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
– Run Completed – When selected, recipients are e-mailed every time the
instrument completes a run.
• Obtain e-mail addresses to receive notifications.
IMPORTANT! Separate addresses with a comma (,).
• Contact your systems administrator or information technology department if you
need:
– E-mail addresses for users who will receive notifications
– A network address for a simple mail transfer protocol (SMTP) server on the LAN
– A user name and password for the server, if required for access
– The Secure Sockets Layer (SSL) setting of the server (on or off)
Start the Run
Start the run according to the layout of your StepOne or StepOnePlus system:
Colocated
Startup
Perform this procedure if your computer is directly connected to your StepOne or
StepOnePlus instrument by the yellow cable.
1. In the StepOne software, click Run in the navigation pane.
2. Click START RUN .
Layout Description See…
Colocated The yellow cable connects the computer to the
instrument
“Colocated Startup”
below
Standalone • The computer and the instrument are not
connected, or
• The computer and the instrument are
connected to the same network.
“Standalone Startup”
on page 66
1
2

Chapter 4 Run the Experiment
Start the Run
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
66
Notes
Standalone
Startup
Perform these procedures if your computer and StepOne or StepOnePlus instrument are
not directly connected by the yellow cable. Start with:
• “Send the Experiment to the Instrument Over the Network” on page 66 if your
computer and instrument are connected to the same network.
or
• “Transfer the Experiment to the Instrument Using a USB Drive” on page 66 if your
computer and instrument are not connected to the same network.
Send the Experiment to the Instrument Over the Network
1. In the StepOne software, click Send Experiment to Instrument.
2. In the Send Experiment to Instrument dialog box:
a. Click Browse, navigate to the example experiment file, then click Open.
b. Select the instrument to receive the experiment file.
Note: If your instrument is not listed, set up the instrument for monitoring as
explained in the Applied Biosystems StepOne
™
and StepOnePlus
™
Real-Time
PCR Systems Installation, Networking, and Maintenance Guide.
c. Click Send Experiment to send the experiment to the your instrument over the
network.
3. When prompted, click OK to close the confirmation.
4. Go to “Start the Instrument Run Using the Touchscreen” on page 67.
Transfer the Experiment to the Instrument Using a USB Drive
1. Connect the USB drive to one of the USB ports on the computer.
2. In the StepOne software, select SaveSave As.
3. In the Save dialog box, navigate to the USB drive, then click Save.
2a
2b
2c

Chapter 4 Run the Experiment
Start the Run
67
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
4. Remove the USB drive from your computer, then connect it to the USB port of your
StepOne or StepOnePlus instrument.
5. Go to “Start the Instrument Run Using the Touchscreen” below.
Start the Instrument Run Using the Touchscreen
1. Touch the StepOne or StepOnePlus instrument touchscreen to awaken it.
Note: If the touchscreen is not at the Main Menu screen, touch .
2. Wait for the USB sign to appear on the touchscreen.
3. In the Main Menu screen, touch Browse/New Experiments.
4. In the Browse screen, touch Folders.
5. In the Choose an Experiment Folder screen:
• Touch USB if you transferred the experiment on a USB drive.
• Touch Default if you sent the experiment over a network connection.
6. Before starting the run, save the example experiment to your instrument:
a. In the Browse screen, touch the example experiment name, then touch Copy.
b. In the Save Experiment screen, navigate to a destination folder, then click
Save & Exit.
7. In the Browse screen, touch the example experiment name, then touch
Start Run.

Chapter 4 Run the Experiment
Start the Run
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
68
Notes
8. In the Run Parameters screen:
a. Touch the Reaction Volume field, use the keypad to enter the reaction volume
for the example experiment, then touch Done.
b. Touch Start Run Now.
4
6a
7
8b
8a

Chapter 4 Run the Experiment
Monitor the Run
69
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Monitor the Run
Monitor the run according to the layout of your StepOne or StepOnePlus system:
Colocated
Monitoring
If your computer is directly connected to your StepOne or StepOnePlus instrument by the
yellow cable, you can view the progress of the run in realtime as described below. During
the run, periodically view all three plots available from the StepOne software for
potential problems.
Layout Description See…
Colocated The yellow cable connects the computer to the
instrument.
“Colocated Monitoring”
below
Standalone
(Networked)
The computer and the instrument are
connected to the same network.
“Remote Monitor” on
page 73
Standalone
(Basic)
The computer and the instrument are not
connected.
“Standalone Monitoring”
on page 75
# To… Action
A Stop the run 1. In the StepOne software, click STOP RUN.
2. In the Stop Run dialog, click one of the following:
– Stop Immediately to stop the run immediately.
– Stop after Current Cycle/Hold to stop the run after the
current cycle or hold.
– Cancel to continue the run.
B View amplification data
in realtime
Select Amplification Plot.
See “About the Amplification Plot Screen” on page 70.
C View temperature data
for the run in realtime
Select Temperature Plot.
See “About the Temperature Plot Screen” on page 71.
D View progress of the
run in the Run Method
screen
Select Run Method.
See “About the Run Method Screen” on page 72.
E Enable/disable the
Notification Settings
Select or deselect Enable Notifications.
See “(Optional) Enable the Notification Settings” on page 63.

Chapter 4 Run the Experiment
Monitor the Run
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
70
Notes
About the Amplification Plot Screen
The Amplification Plot screen allows you to view sample amplification as your
instrument collects fluorescence data during a run. If a method is set up to collect real-
time data, the Amplification Plot screen displays the data for the wells selected in the
View Plate Layout tab. The plot contrasts normalized dye fluorescence (ΔRn) and cycle
number. The figure below shows the Amplification Plot screen as it appears during the
example experiment.
To view data in the Amplification Plot screen, select the wells that you want to view in
the View Plate Layout tab.
The Amplification Plot screen is useful for identifying and examining abnormal
amplification. Abnormal amplification can include the following:
• Increased fluorescence in negative control wells.
C
B
D
A
E

Chapter 4 Run the Experiment
Monitor the Run
71
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
• Absence of detectable fluorescence at an expected cycle (determined from previous
similar experiments run using the same reagents under the same conditions).
If you notice abnormal amplification or a complete absence of signal, troubleshoot the
error as explained in the StepOne Software Help (click or press F1).
About the Temperature Plot Screen
During a run, the Temperature Plot screen displays the temperatures of the sample
block(s), the heated cover, and samples (calculated) in realtime. The figure below shows
the Temperature Plot screen as it appears during the example experiment.
The Temperature Plot screen can be useful for identifying hardware failures. When
monitoring the Temperature Plot screen, observe the Sample and Block plots for
abnormal behavior.
To … A ct i o n
A Add/remove temperature plots Select Cover or Sample Block to
toggle the presence of the
associated data in the plot.
B Change the time displayed by plot From the View dropdown menu,
select the amount of time to display
in the plot.
C Display a fixed time window during the instrument
run
If the entire plot does not fit in the screen, the
screen is not updated as the run progresses. For
example, if you select 10 minutes from the View
dropdown menu, the plot will show data for 10
minutes. If the run lasts more than 10 minutes:
• The plot updates as the run progresses with
Fixed View deselected.
• The plot does not update as the run progresses
with Fixed View selected.
Select Fixed View.
A
B
C

Chapter 4 Run the Experiment
Monitor the Run
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
72
Notes
• In general, the Sample and Block plots should mirror each other approximately. A
significant deviation of the plots may indicate a problem.
• The Cover plot should maintain the constant temperature specified in the method. A
departure from the
constant temperature may indicate a problem.
If you notice an abnormal temperature plot, troubleshoot the error as explained in the
StepOne Software Help (click or press F1).
Note: The Sample temperature displayed in the Current Temperatures group is an
estimated value.
About the Run Method Screen
The Run Method screen displays the run method selected for the run in progress. The
software updates the Run Status field throughout the run. The figure below shows the
Run Method screen as it appears in the example experiment.
If an alert appears, click the error for more information and troubleshoot the problem as
explained in the StepOne Software Help (click or press F1).
To … A ct i o n
A Change the number of cycles In the Adjust # of Cycles field, enter the number of
cycles to apply to the Cycling Stage.
B Add a melt curve stage to the
end of the run
Select Add Melt Curve Stage to End.
C Add a Hold stage to the end of
the run
Select Add Holding Stage to End.
D Apply your changes Click Send to Instrument.
A
C
D
B

Chapter 4 Run the Experiment
Monitor the Run
73
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Remote Monitor
If your StepOne or StepOnePlus instrument is connected to a network, you can use the
Remote Monitor in the StepOne software to view the progress of the run in realtime from
any computer on the network.
IMPORTANT! Networked computers cannot control the StepOne or StepOnePlus
instrument, only monitor it.
To monitor your instrument remotely:
1. In the StepOne software, select InstrumentRemote Monitor.
2. In the navigation pane, select your instrument.
If the navigation pane does not list your instrument:
a. Click Add Instrument.
b. Enter a name for the instrument profile within the Remote Monitor.
Note: Enter any name that helps you identify the instrument. The profile name
you enter will be displayed in the Remote Monitor and in the instrument
dropdown menus when you send experiments, download experiments, or
monitor instruments.
c. In the Instrument Name, Host Name, or IP Address field:
• If you know the host name, enter the host name.
• If you do not know the host name, enter the instrument name or IP
address.
Note: The instrument name and IP address are displayed on the instrument
touchscreen. Go to Settings MenuAdmin MenuSet Instrument Name
or Set IP Address. Contact your systems administrator or information
technology department for the host name.
d. Click Save & Exit.
Note: For more information on configuring the StepOne or StepOnePlus instrument
for network use or for the Remote Monitor feature, refer to the Applied Biosystems
StepOne
™
and StepOnePlus
™
Real-Time PCR Systems Installation, Networking,
and Maintenance Guide.
2c
2b
2d

Chapter 4 Run the Experiment
Monitor the Run
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
74
Notes
3. Click Start monitoring the instrument for your instrument. It may take
several minutes for the instrument to send the information to your computer.
4. View data as described below.
2
1
3
# To… Action
A View amplification data
in realtime
Click Amplification Plot.
See “About the Amplification Plot Screen” on page 70.
B View temperature data
for the run in realtime
Click Temperature Plot.
See “About the Temperature Plot Screen” on page 71.
C View progress of the
run in the Run Method
screen
Click Run Method.
See “About the Run Method Screen” on page 72.
D Enable/disable the
Notification Settings
Select or deselect Enable Notifications.
See “(Optional) Enable the Notification Settings” on page 63.

Chapter 4 Run the Experiment
Monitor the Run
75
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Standalone
Monitoring
If you started the run from your StepOne or StepOnePlus instrument, you can view the
progress of the run from the touchscreen. The Run Method screen displays the method
for the experiment and highlights the thermal profile steps as the instrument performs
them.
A B C
D
# To… Action
A Add a melt curve stage
to the run
Touch Add Melt Curve, then touch OK.
B Display the time
remaining in the run
Touch Display Experiment Time, then touch to return
to the Run Method screen.
C Stop the run Touch STOP, then touch:
• Stop to stop the run after the instrument completes the
current cycle or hold.
• Abort to stop the run immediately.
• to continue the run with no changes.
D View experiment
information
Touch View Experiment Information, then touch to
return to the Run Method screen.
E View the Error Log Touch the status bar to display the error log.
D
A
E
B
C

Chapter 4 Run the Experiment
Unload the Instrument and Transfer the Data
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
76
Notes
Unload the Instrument and Transfer the Data
When your StepOne or StepOnePlus instrument displays the Main Menu screen, unload
the reaction plate from the instrument and transfer the experiment data to the computer
for analysis.
Unload the
Reaction Plate
PHYSICAL INJURY HAZARD. During instrument operation, the
temperature of the sample block(s) can exceed 100 °C. Keep your hands away until the
sample block(s) reaches room temperature.
Note: When the StepOne or StepOnePlus instrument completes a run, the system saves
the details of the run to the run history, which remains present in the system until the
instrument completes another run.
1. When the Run Report screen appears in the StepOne or StepOnePlus instrument
touchscreen, touch .
2. Open the instrument drawer.
3. Remove the reaction plate from the sample block(s).
4. Carefully close the instrument drawer.
Select a Data
Transfer Method
Transfer the experiment to your computer for analysis according to the layout of your
StepOne or StepOnePlus system:
Layout Description See…
Colocated The yellow cable connects the computer and
the instrument.
“Colocated Data Transfer”
below
Standalone
(Networked)
The computer and the instrument are
connected to the same network.
“Remote Data Transfer” on
page 77
Standalone
(Basic)
The computer and the instrument are not
connected.
“Standalone
Data Transfer” on page 78

Chapter 4 Run the Experiment
Unload the Instrument and Transfer the Data
77
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Colocated
Data Transfer
If your computer is directly connected to your StepOne or StepOnePlus instrument by the
yellow cable, no action is necessary. The StepOne software automatically transfers the
experiment data from the instrument to the computer after the run.
Note: In a colocated layout, you can start the run from the computer or from the
instrument touchscreen. However, the StepOne software only transfers the experiment
data automatically when a run is started from the computer (see “Colocated Startup” on
page 65).
Remote
Data Transfer
If your computer and StepOne or StepOnePlus instrument are connected to the same
Ethernet network, download the experiment from the instrument over the network:
1. In the StepOne software, click Download Experiment from Instrument to
open the Download Experiment from Instrument dialog box.
2. From the Select Instrument dropdown menu, select your instrument.
3. From the Experiment dropdown menu, select the example experiment file.
4. In the Download File To field:
a. Click Browse.
b. Navigate to:
<drive>:\Applied Biosystems\<software name>\experiments\
where:
<drive> is the computer hard drive on which the StepOne software is installed.
The default installation drive for the software is the D drive.
<software name> is the current version of the StepOne software.
c. Click Select.
5. Click Download Experiment to download the example experiment file from your
instrument to your computer over the network.
6. When prompted, click OK to close the confirmation.
2
4
5
3
6

Chapter 4 Run the Experiment
Unload the Instrument and Transfer the Data
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
78
Notes
Standalone
Data Transfer
If your computer is not connected to your StepOne or StepOnePlus instrument, use the
USB drive to transfer the experiment from the instrument to the computer:
1. If not already connected to the instrument, connect a USB drive to the USB port.
o
2. Touch the StepOne or StepOnePlus instrument touchscreen to awaken it.
Note: If the touchscreen is not at the Main Menu screen, touch .
3. Wait for the USB sign to appear on the touchscreen.
4. In the Main Menu, touch Collect Results to save the data to the USB drive.
Note: If your instrument cannot find the USB drive, remove the USB drive, then try
again. If the instrument still does not recognize the USB drive, try another USB
drive.
5. When prompted that the data has been transferred successfully, touch OK.
6. Remove the USB drive from your instrument, then connect it to one of the USB
ports on your computer.
7. In the computer desktop, use the Windows explorer open the USB drive.

Chapter 4 Run the Experiment
Unload the Instrument and Transfer the Data
79
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
8. Copy the example experiment file to:
<drive>:\Applied Biosystems\<software name>\experiments\
where:
• <drive> is the computer hard drive on which the StepOne software is installed.
The default installation drive for the software is the D drive.
• <software name> is the current version of the StepOne software.

Chapter 4 Run the Experiment
Unload the Instrument and Transfer the Data
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
80
Notes

Chapter 5
81
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Analyze the Experiment
This chapter covers:
■ Chapter Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 82
■ Section 5.1 Review Results. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83
■ Section 5.2 Troubleshoot (If Needed). . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 103
Note: For more information about any of the topics discussed in this guide, access the
Help from within Applied Biosystems StepOne
™
Real-Time PCR Software v2.0 by
pressing F1, clicking in the toolbar, or selecting HelpStepOne Software Help.

Chapter 5 Analyze the Experiment
Chapter Overview
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
82
Notes
Chapter Overview
The StepOne
™
software analyzes the data using the standard curve quantitation method.
Section 1 of this chapter explains how to review the analyzed data using several of the
analysis screens and how to publish the data. If you receive questionable results,
Section 2 of this chapter explains how to perform some troubleshooting steps.
Example
Experiment
Workflow
The workflow for analyzing the example experiment provided with this getting started
guide is shown below.
Design the Experiment (Chapter 2)
Start Experiment
Prepare the Reactions (Chapter 3)
Run the Experiment (Chapter 4)
End Experiment
Analyze the Experiment (Chapter 5)
Section 1, Review Results:
1. Analyze.
2. View the standard curve.
3. View the amplification plot.
4. View the results in a table.
5. Publish the data.
Section 2, Troubleshoot (If Needed):
1. View the analysis settings; adjust the baseline/threshold.
2. View the quality summary.
3. Omit wells.
4. View the multicomponent plot.
5. View the raw data plot.

Chapter 5 Analyze the Experiment
Section 5.1 Review Results
83
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Section 5.1 Review Results
This section covers:
■ Analyze the Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84
■ View the Standard Curve . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 90
■ View the Amplification Plot . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 92
■ View the Well Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 99
■ Publish the Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 102

Chapter 5 Analyze the Experiment
Analyze the Experiment
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
84
Notes
Analyze the Experiment
The StepOne software analyzes the experiment and displays results in the analysis
screens (for example, the Amplification Plot screen, QC Summary screen, and so on).
About the
Example
Experiment
For the standard curve example experiment, use the data file that installs with the
StepOne software. The data file was created with the same design parameters provided in
Chapter 2, then run and analyzed on a StepOne
™
instrument. You can find the data file
for the example experiment on your computer:
<drive>:\Applied Biosystems\<software name>\experiments\examples\
Standard Curve Example.eds
where:
• <drive> is the computer hard drive on which the StepOne software is installed. The
default installation drive for the software is the D drive.
• <software name> is the current version of the StepOne software.
Analyze the
Example
Experiment
1. Double-click (StepOne software shortcut) or select
StartAll ProgramsApplied BiosystemsStepOne Software
<software name>
where <software name> is the current version of the StepOne software.
2. From the Home screen, click Open.
3. In the Open dialog box, navigate to the examples folder:
<drive>:\Applied Biosystems\<software name>\experiments\examples
4. Double-click Standard Curve Example to open the example experiment data file.
Note: The examples folder contains several data files; be sure to select Standard
Curve Example. For information on the other data files, see “Data Files in the
Examples Folder” on page 12.

Chapter 5 Analyze the Experiment
Analyze the Experiment
85
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
5. Click Analyze. The software analyzes the data using the default analysis settings.
6. See “Software Elements” below and “Navigation Tips” on page 88 for information
on navigating within the analysis screens.
Guidelines
When you analyze your own standard curve experiment:
• Immediately after a run, the StepOne software automatically analyzes the data using
the default analysis settings, then displays the Amplification Plot screen on your
computer.
• To reanalyze the data, select all the wells in the plate layout, then click Analyze.
Software
Elements
The StepOne software elements for the analysis screens are illustrated below.
1. Menu bar – Displays the menus available in the software:
•File
•Edit
• Instrument
•Analysis
• Tools
•Help
2
4
3
5

Chapter 5 Analyze the Experiment
Analyze the Experiment
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
86
Notes
2. Toolbar – Displays the tools available in the software:
• New Experiment
•Open
•Save
•Close
• Send Experiment to Instrument
• Download Experiment from Instrument
• Export
• Print Report
3. Experiment header - Displays the experiment name, experiment type, and reagents
for the open experiment.
4. Experiment Menu pane – Provides links to the following software screens:
• Setup screens
•Run screens
• Analysis screens:
• Amplification Plot
•Standard Curve
• Multicomponent Plot
•Raw Data Plot
• QC Summary
• Multiple Plots View
5. Plot pane – Displays the selected analysis screen for the open experiment.
6. View tabs – Displays the plate layout or Well Table for the open experiment
7. Experiment tab(s) – Displays a tab for each open experiment.

Chapter 5 Analyze the Experiment
Analyze the Experiment
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
88
Notes
Navigation Tips
How to Select Wells
To display specific wells in the analysis screens, select the wells in the View Plate
Layout tab as follows:
1. To select wells of a specific type, use the Select Wells With dropdown menus:
Select Sample, Target, or Task, then select the sample, target, or task name.
2. To select a single well, click the well in the plate layout.
3. To select multiple wells, click and drag over the desired wells, press CTRL+click,
or press Shift+click in the plate layout.
4. To select all 48 wells, click the upper left corner of the plate layout.
1
4

Chapter 5 Analyze the Experiment
Analyze the Experiment
89
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
How To Display Multiple Plots
Use the Multiple Plots screen to display up to four plots simultaneously. To navigate
within the Multiple Plots screen:
1. From the Experiment Menu pane, select Analysis Multiple Plots View.
2. To display four plots, click Show plots in a 2 ✕ 2 matrix.
3. To display two plots in rows, click Show plots in two rows.
4. To display two plots in columns, click Show plots in two columns.
5. To display a specific plot, select the plot from the dropdown menu above each plot
display.
4
2 35

Chapter 5 Analyze the Experiment
View the Standard Curve
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
90
Notes
View the Standard Curve
The Standard Curve screen displays the standard curve for samples designated as
standards. The StepOne software calculates the quantity of an unknown target from the
standard curve.
About the
Example
Experiment
In the standard curve example experiment, you review the Standard Curve screen for the
following regression coefficient values:
• Slope/amplification efficiency
•R
2
value (correlation coefficient)
•C
T
values
View the
Standard Curve
1. From the Experiment Menu pane, select Analysis Standard Curve.
Note: If no data are displayed, click Analyze.
2. Display all 48 wells in the Standard Curve screen by clicking the upper left corner
of the plate layout in the View Plate Layout tab.
3. From the Target dropdown menu, select All.
4. From the Plot Color dropdown menu, select Default.
5. Click Show a legend for the plot (default).
Note: This is a toggle button. When the legend is displayed, the button changes to
Hide the plot legend.
6. View the values displayed below the standard curve. In the example experiment, the
values for the target (RNase P) fall within the acceptable ranges:
– The slope is −3.477.
–The R
2
value is 0.999.
– The amplification efficiency (Eff%) is 93.912%.
7. Check that all samples are within the standard curve. In the example experiment, all
samples (blue dots) are within the standard curve (red dots).

Chapter 5 Analyze the Experiment
View the Standard Curve
91
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
8. Check the C
T
values:
a. Click the View Well Table tab.
b. From the Group By dropdown menu, select Replicate.
c. Look at the values in the C
T
column. In the example experiment, the C
T
values
fall within the expected range (>8 and <35).
6
3 4
5
7
8b
8a
8c

Chapter 5 Analyze the Experiment
View the Amplification Plot
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
92
Notes
Analysis
Guidelines
When you analyze your own standard curve experiment, look for:
• Slope/amplification efficiency values – The amplification efficiency is calculated
using the slope of the regression line in the standard curve. A slope close to −3.3
indicates optimal, 100% PCR amplification efficiency. Factors that affect
amplification efficiency:
– Range of standard quantities – For more accurate and precise efficiency
measurements, use a broad range of standard quantities, 5 to 6 logs
(10
5
to 10
6
fold).
– Number of standard replicates – For more accurate efficiency measurements,
include replicates to decrease the effects of pipetting inaccuracies.
– PCR inhibitors – PCR inhibitors in the reaction can reduce amplification
efficiency.
• R
2
values (correlation coefficient) – The R
2
value is a measure of the closeness of
fit between the regression line and the individual C
T
data points of the standard
reactions. A value of 1.00 indicates a perfect fit between the regression line and the
data points. An R
2
value >0.99 is desirable.
• C
T
values – The threshold cycle (C
T
) is the PCR cycle number at which the
fluorescence level meets the threshold. A C
T
value >8 and <35 is desirable. A C
T
value <8 indicates that there is too much template in the reaction. A C
T
value >35
indicates a low amount of target in the reaction; for C
T
values >35, expect a higher
standard deviation.
If your experiment does not meet the guidelines above, troubleshoot as follows:
• Omit wells (see “Omit Wells from the Analysis” on page 108).
Or
• Rerun the experiment.
For More
Information
For more information on:
• The Standard Curve screen, access the StepOne Software Help by clicking or
pressing F1.
• Amplification efficiency, refer to the Amplification Efficiency of TaqMan
®
Gene
Expression Assays Application Note.
View the Amplification Plot
The Amplification Plot screen displays amplification of all samples in the selected wells.
There are three plots available:
• ΔRn vs Cycle – ΔRn is the magnitude of normalized fluorescence signal generated
by the reporter at each cycle during the PCR amplification. This plot displays ΔRn
as a function of cycle number. You can use this plot to identify and examine
irregular amplification and to view threshold and baseline values for the run.

Chapter 5 Analyze the Experiment
View the Amplification Plot
93
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
• Rn vs Cycle – Rn is the fluorescence signal from the reporter dye normalized to the
fluorescence signal from the passive reference. This plot displays Rn as a function
of cycle number. You can use this plot to identify and examine irregular
amplification.
• C
T
vs Well – C
T
is the PCR cycle number at which the fluorescence meets the
threshold in the amplification plot. This plot displays C
T
as a function of well
position. You can use this plot to locate outlying amplification (outliers).
Each plot can be viewed as the following graph types: linear or log10.
About the
Example
Experiment
In the standard curve example experiment, you review the target in the Amplification
Plot screen for:
• Correct baseline and threshold values
•Outliers
View the
Amplification Plot
1. From the Experiment Menu pane, select Analysis Amplification Plot.
Note: If no data are displayed, click Analyze.
2. Display the RNase P wells in the Amplification Plot screen:
a. Click the View Plate Layout tab.
b. From the Select Wells With dropdown menus, select Target, then RNase P.
3. In the Amplification Plot screen:
a. From the Plot Type dropdown menu, select ΔRn vs Cycle (default).
b. From the Plot Color dropdown menu, select Wel l (default).
2a 2b

Chapter 5 Analyze the Experiment
View the Amplification Plot
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
94
Notes
c. Click Show a legend for the plot (default).
Note: This is a toggle button. When the legend is displayed, the button changes
to Hide the plot legend.
4. View the baseline values:
a. From the Graph Type dropdown menu, select Linear.
b. Select the Baseline checkbox to show the start cycle and end cycle.
c. Verify that the baseline is set correctly: The end cycle should be set a few
cycles before the cycle number where significant fluorescent signal is detected.
In the example experiment, the baseline is set correctly.
5. View the threshold values:
a. From the Graph Type dropdown menu, select Log.
b. From the Target dropdown menu, select RNase P.
c. Select the Threshold checkbox to show the threshold.
d. Verify that the threshold is set correctly. In the example experiment, the
threshold is in the exponential phase.
4b
3c
3a 4a 3b

Chapter 5 Analyze the Experiment
View the Amplification Plot
95
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
6. Locate any outliers:
a. From the Plot Type dropdown menu, select C
T
vs Well.
b. Look for outliers from the amplification plot. In the example experiment, there
are no outliers for RNase P.
5c
5a
5b
6a

Chapter 5 Analyze the Experiment
View the Amplification Plot
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
96
Notes
Analysis
Guidelines
When you analyze your own standard curve experiment, look for:
•Outliers
• A typical amplification plot – The StepOne software automatically calculates
baseline and threshold values based on the assumption that the data exhibit a typical
amplification plot. A typical amplification plot has four distinct sections:
a. Plateau phase
b. Linear phase
c. Exponential (geometric phase)
d. Baseline
IMPORTANT! Experimental error (such as contamination or pipetting errors) can
produce atypical amplification curves that can result in incorrect baseline and
threshold value calculations by the StepOne software. Therefore, Applied
Biosystems recommends that you examine the Amplification Plot screen and review
the assigned baseline and threshold values for each well after analysis completes.
• Correct baseline and threshold values – See the threshold examples on page 97 and
the baseline examples on page 98.
Cycle
ΔRn
a
b
c
d
Threshold

Chapter 5 Analyze the Experiment
View the Amplification Plot
97
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Threshold Set Correctly
The threshold is set in the exponential phase of the
amplification curve.
Threshold settings above or below the optimum
increase the standard deviation of the replicate
groups.
Threshold Set Too Low
The threshold is set below the exponential phase of
the amplification curve. The standard deviation is
significantly higher than that for a plot where the
threshold is set correctly. Drag the threshold bar up
into the exponential phase of the curve.
Threshold Set Too High
The threshold is set above the exponential phase of
the amplification curve. The standard deviation is
significantly higher than that for a plot where the
threshold is set correctly. Drag the threshold bar
down into the exponential phase of the curve.
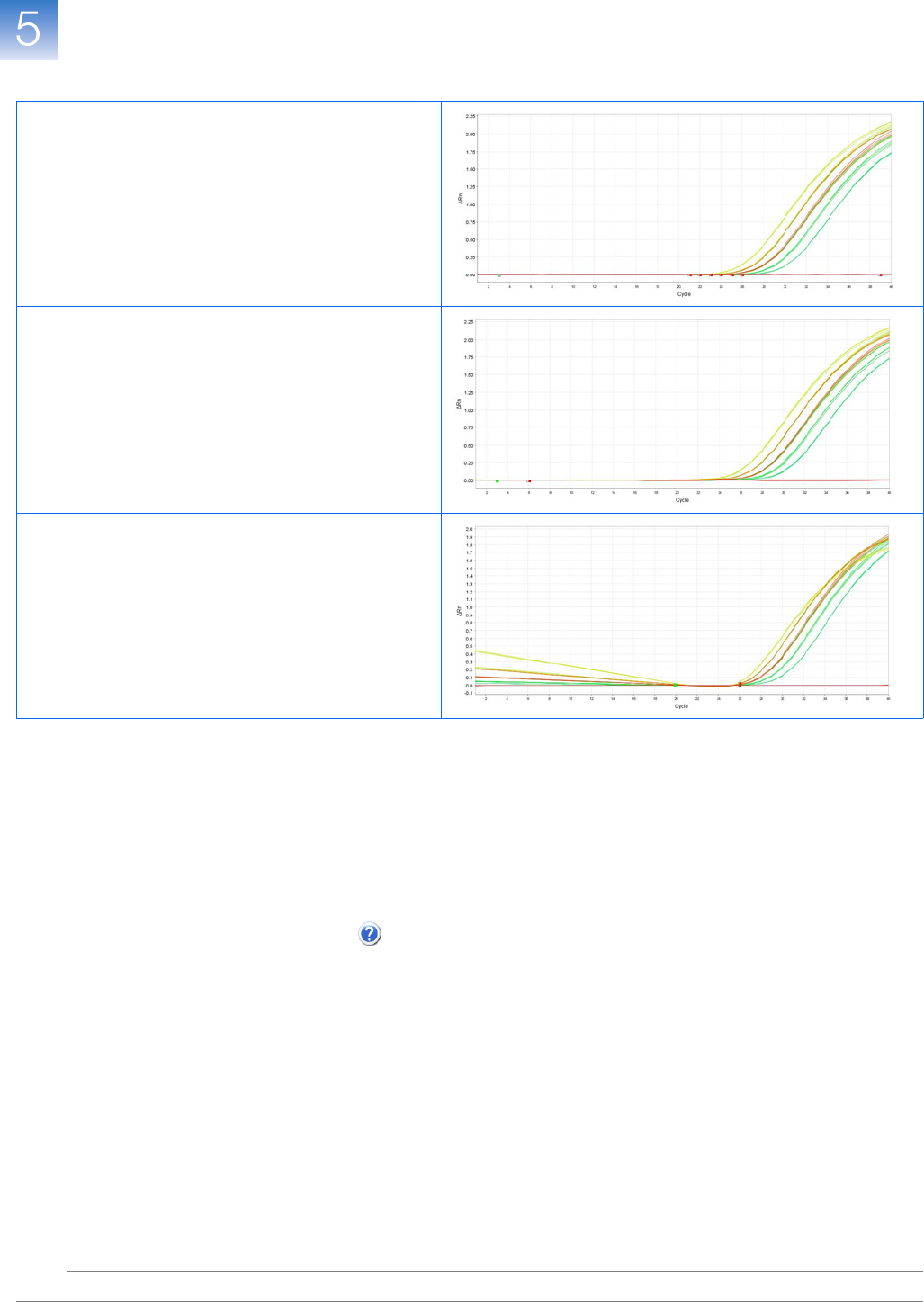
Chapter 5 Analyze the Experiment
View the Amplification Plot
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
98
Notes
If your experiment does not meet the guidelines above, troubleshoot as follows:
• Omit wells (see “Omit Wells from the Analysis” on page 108).
or
• Manually adjust the baseline and/or threshold (see “View the Analysis Settings” on
page 104).
For More
Information
For more information on the Amplification Plot screen, access the StepOne Software
Help by clicking or pressing F1.
Baseline Set Correctly
The amplification curve begins after the maximum
baseline.
Baseline Set Too Low
The amplification curve begins too far to the right
of the maximum baseline. Increase the End Cycle
value.
Baseline Set Too High
The amplification curve begins before the
maximum baseline. Decrease the End Cycle value.

Chapter 5 Analyze the Experiment
View the Well Table
99
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
View the Well Table
The Well Table displays data for each well in the reaction plate, including:
• The sample name, target name, task, and dyes
• The calculated threshold cycle (C
T
), normalized fluorescence (Rn), and quantity
values
• Comments
•Flags
About the
Example
Experiment
In the standard curve example experiment, you review the Well Table for:
• Quantity values
•Flags
•C
T
values (including C
T
standard deviation)
View the Well
Table
1. From the Experiment Menu pane, select Analysis, then select the View Well Table
tab.
Note: If no data are displayed, click Analyze.
2. Use the Group By dropdown menu to group wells by a specific category. For the
example experiment, group the wells by replicate, flag, or C
T
value.
Note: You can select only one category at a time.
a. From the Group By dropdown menu, select Replicate. The software groups the
replicate wells: negative controls, standards, and samples. In the example
experiment, note that the quantity values within each replicate group are
similar.
Note: In the example experiment, the Quantity, Quantity Mean, and Quantity
SD columns have been moved from their default locations to the beginning of
the Well Table. To move a column, click and drag on the column heading.

Chapter 5 Analyze the Experiment
View the Well Table
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
100
Notes
b. From the Group By dropdown menu, select Flag. The software groups the
flagged and unflagged wells. In the example experiment, there are no flagged
wells.
2a
2b

Chapter 5 Analyze the Experiment
View the Well Table
101
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
c. From the Group By dropdown menu, select C
T
. The software groups the wells
by C
T
value: low, medium, high, and undetermined. In the example experiment,
the C
T
values are within the expected range (>8 and <35).
Analysis
Guidelines
When you analyze your own standard curve experiment, group the wells by:
• Replicate – The software groups the wells by replicate: negative controls, standards,
and samples. Look in the Quantity columns to make sure the quantity values for
each replicate group are similar. This indicates tight precision.
• Flag – The software groups the flagged and unflagged wells. A flag indicates that
the software has found an error in the flagged well. For a description of the StepOne
software flags, see “View the QC Summary” on page 106.
• C
T
– The threshold cycle (C
T
) is the PCR cycle number at which the fluorescence
level meets the threshold. A C
T
value >8 and <35 is desirable. A C
T
value <8
indicates that there is too much template in the reaction. A C
T
value >35 indicates a
low amount of target in the reaction; for C
T
values >35, expect a higher standard
deviation.
For More
Information
For more information on the Well Table, access the StepOne Software Help by
clicking or pressing F1.
2c

Chapter 5 Analyze the Experiment
Publish the Data
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
102
Notes
Publish the Data
You can publish the experiment data in several ways:
• Save the plot as an image file
• Print the plot
• Print the plate layout
• Create slides
• Print a report
• Export data
For information on performing these procedures, access the StepOne Software Help by
clicking or pressing F1.

Chapter 5 Analyze the Experiment
Section 5.2 Troubleshoot (If Needed)
103
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Section 5.2 Troubleshoot (If Needed)
This section covers:
■ View the Analysis Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 104
■ View the QC Summary. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 106
■ Omit Wells from the Analysis. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 108
■ View the Multicomponent Plot. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 109
■ View the Raw Data Plot . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 112

Chapter 5 Analyze the Experiment
View the Analysis Settings
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
104
Notes
View the Analysis Settings
The Analysis Settings dialog box displays the analysis settings for the threshold cycle
(C
T
), flags, and advanced options. If the default analysis settings in the StepOne software
are not suitable for your experiment, you can change the settings in the Analysis Settings
dialog box, then reanalyze your experiment.
About the
Example
Experiment
In the standard curve example experiment, the default analysis settings are used without
changes.
View the Analysis
Settings
1. From the Experiment Menu pane, select Analysis.
2. Click Analysis Settings to open the Analysis Settings dialog box.
3. In the example experiment, the default analysis settings are used for each tab:
•C
T
Settings
• Flag Settings
• Advanced Settings

Chapter 5 Analyze the Experiment
View the Analysis Settings
105
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Analysis
Guidelines
Unless you have already determined the optimal settings for your experiment, use the
default analysis settings in the StepOne software. If the default settings are not suitable
for your experiment, you can change the:
• C
T
Settings – Use this tab to manually set the threshold and baseline. When
manually setting the threshold and baseline, Applied Biosystems recommends the
following:
• Flag Settings – Use this tab to:
– Adjust the sensitivity so that more wells or fewer wells are flagged.
– Change the flags that are applied by the StepOne software.
• Advanced Settings – Use this tab to change baseline settings well by well.
For More
Information
For more information on the analysis settings, access the StepOne Software Help by
pressing F1 when the Analysis Settings dialog box is open.
Setting Recommendation
Threshold Enter a value for the threshold so that the threshold is:
• Above the background.
• Below the plateau and linear regions of the amplification curve.
• Within the exponential phase of the amplification curve.
Baseline Select the Start Cycle and End Cycle values so that the baseline
ends before significant fluorescent signal is detected.

Chapter 5 Analyze the Experiment
View the QC Summary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
106
Notes
View the QC Summary
The QC Summary screen displays a list of the StepOne software flags, and includes the
flag frequency and location for the open experiment.
About the
Example
Experiment
In the standard curve example experiment, you review the QC Summary screen for any
flags triggered by the experiment data. In the example experiment, no flags have been
triggered.
View the QC
Summary
1. From the Experiment Menu pane, select Analysis QC Summary.
Note: If no data are displayed, click Analyze.
2. Review the Flags Summary. In the example experiment, there are 0 flagged wells.
3. In the Flag Details table, look in the Frequency and Wells columns to determine
which flags appear in the experiment. In the example experiment, the Frequency
column displays 0 for all flags.
Note: A 0 displayed in the Frequency column indicates that the flag does not appear
in the experiment.
4. (Optional) Click each flag row to display detailed information about the flag.
2
3
4

Chapter 5 Analyze the Experiment
View the QC Summary
107
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Possible Flags
For standard curve experiments, the flags listed below may be triggered by the
experiment data.
If a flag does not appear in the experiment, its frequency is 0. If the frequency is >0, the
flag appears somewhere in the experiment; the well position is listed in the Wells
column.
Analysis
Guidelines
When you analyze your own standard curve experiment:
• Click each flag in the Flag Details table with a frequency >0 to display detailed
information about the flag. If needed, click the troubleshooting link to view
information on correcting the flag.
• You can change the flag settings:
– Adjust the sensitivity so that more wells or fewer wells are flagged.
– Change the flags that are applied by the StepOne software.
For More
Information
For more information on the QC Summary screen or on flag settings, access the StepOne
Software Help by clicking or pressing F1.
Flag Description
AMPNC Amplification in negative control
BADROX Bad passive reference signal
BLFAIL Baseline algorithm failed
CTFAIL C
T
algorithm failed
EXPFAIL Exponential algorithm failed
HIGHSD High standard deviation in replicate group
MTP Multiple Tm peaks
Note: This flag is only displayed if the experiment
includes a melt curve.
NOAMP No amplification
NOISE Noise higher than others in plate
NOSIGNAL No signal in well
OFFSCALE Fluorescence is offscale
OUTLIERRG Outlier in replicate group
SPIKE Noise spikes
THOLDFAIL Thresholding algorithm failed

Chapter 5 Analyze the Experiment
Omit Wells from the Analysis
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
108
Notes
Omit Wells from the Analysis
Experimental error may cause some wells to be amplified insufficiently or not at all.
These wells typically produce C
T
values that differ significantly from the average for the
associated replicate wells. If included in the calculations, these outliers can result in
erroneous measurements; to ensure precision, omit the outliers from the analysis.
About the
Example
Experiment
In the standard curve example experiment, there are no outliers; no wells need to be
removed from analysis.
Omit Wells
1. From the Experiment Menu pane, select Analysis Amplification Plot.
Note: If no data are displayed, click Analyze.
2. In the Amplification Plot screen, select C
T
vs Well from the Plot Type dropdown
menu.
3. Select the View Well Table tab.
4. In the Well Table:
a. From the Group By dropdown menu, select Replicate.
b. Look for any outliers in the replicate group (be sure they are flagged). In the
example experiment, there are no outliers.
4a
3

Chapter 5 Analyze the Experiment
View the Multicomponent Plot
109
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Analysis
Guidelines
When you analyze your own standard curve experiment, carefully view the replicate
groups for outliers. If needed, remove outliers manually using the Well Table:
1. From the Experiment Menu pane, select Analysis Amplification Plot.
Note: If no data are displayed, click Analyze.
2. In the Amplification Plot screen, select C
T
vs Well from the Plot Type dropdown
menu.
3. Select the View Well Table tab.
4. In the Well Table:
a. From the Group By dropdown menu, select Replicate.
b. Look for any outliers in the replicate group (be sure they are flagged).
c. Select the Omit checkbox next to the outlying well(s).
5. Click Analyze to reanalyze the experiment data with the outlying well(s) removed
from the analysis.
For More
Information
For more information on omitting wells from the analysis, access the StepOne Software
Help by clicking or pressing F1. Within the Help, search for the omit well topics:
1. Click the Search tab.
2. Enter omit well.
3. Click List Topics.
4. Double-click the topics you want to review.
View the Multicomponent Plot
The Multicomponent Plot screen displays the complete spectral contribution of each dye
in a selected well over the duration of the PCR run.
About the
Example
Experiment
In the standard curve example experiment, you review the Multicomponent Plot screen
for:
•ROX
™
dye (passive reference)
•FAM
™
dye (reporter)
• Spikes, dips, and/or sudden changes
• Amplification in the negative control wells

Chapter 5 Analyze the Experiment
View the Multicomponent Plot
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
110
Notes
View the
Multicomponent
Plot
1. From the Experiment Menu pane, select Analysis Multicomponent Plot.
Note: If no data are displayed, click Analyze.
2. Display the unknown and standard wells one at a time in the Multicomponent Plot
screen:
a. Click the View Plate Layout tab.
b. Select one well in the plate layout; the well is shown in the Multicomponent
Plot screen.
Note: If you select multiple wells, the Multicomponent Plot screen displays
the data for all selected wells simultaneously.
3. From the Plot Color dropdown menu, select Dye.
4. Click Show a legend for the plot (default).
Note: This is a toggle button. When the legend is displayed, the button changes to
Hide the plot legend.
5. Check the ROX dye signal. In the example experiment, the ROX dye signal remains
constant throughout the PCR process, which indicates typical data.
6. Check the FAM dye signal. In the example experiment, the FAM dye signal
increases throughout the PCR process, which indicates normal amplification.
2b
3 2a4
5
6

Chapter 5 Analyze the Experiment
View the Multicomponent Plot
111
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
7. Select the negative control wells one at time and check for amplification. In the
example experiment, there is no amplification in the negative control wells.
Analysis
Guidelines
When you analyze your own standard curve experiment, look for:
• Passive reference – The passive reference dye fluorescence level should remain
relatively constant throughout the PCR process.
• Reporter dye – The reporter dye fluorescence level should display a flat region
corresponding to the baseline, followed by a rapid rise in fluorescence as the
amplification proceeds.
• Any irregularities in the signal – There should not be any spikes, dips, and/or
sudden changes in the fluorescent signal.
• Negative control wells – There should not be any amplification in the negative
control wells.
For More
Information
For more information on the Multicomponent Plot screen, access the StepOne Software
Help by clicking or pressing F1.
7

Chapter 5 Analyze the Experiment
View the Raw Data Plot
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
112
Notes
View the Raw Data Plot
The Raw Data Plot screen displays the raw fluorescence signal (not normalized) for each
optical filter for the selected wells during each cycle of the real-time PCR.
About the
Example
Experiment
In the standard curve example experiment, you review the Raw Data Plot screen for a
stable increase in signal (no abrupt changes or dips) from the appropriate filter.
View the Raw
Data Plot
1. From the Experiment Menu pane, select Analysis Raw Data Plot.
Note: If no data are displayed, click Analyze.
2. Display all 48 wells in the Raw Data Plot screen by clicking the upper left corner of
the plate layout in the View Plate Layout tab.
3. Click Show a legend for the plot (default).
Note: This is a toggle button. When the legend is displayed, the button changes to
Hide the plot legend.
Note: The legend displays the color code for each row of the reaction plate. In the
example shown below, Row A is red, Row B is yellow/green, Row C is green, and
so on.
4. Click and drag the Show Cycle pointer from cycle 1 to cycle 40. In the example
experiment, there is a stable increase in signal from filter 1, which corresponds to
the FAM
™
dye filter.

Chapter 5 Analyze the Experiment
View the Raw Data Plot
113
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
The filters are:
Analysis
Guidelines
When you analyze your own standard curve experiment, look for the following in each
filter:
• Characteristic signal growth
• No abrupt changes or dips
For More
Information
For more information on the Raw Data Plot screen, access the StepOne Software Help by
clicking or pressing F1.
3
4
StepOne system StepOnePlus system
Filter Dye Filter Dye
1FAM
™
dye 1 FAM
™
dye
SYBR
®
Green dye SYBR
®
Green dye
2JOE
™
dye 2 JOE
™
dye
VIC
®
dye VIC
®
dye
3ROX
™
dye 3 TAMRA
™
dye
NED
™
dye
4ROX
™
dye

Chapter 5 Analyze the Experiment
View the Raw Data Plot
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
114
Notes

Appendix A
115
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Alternate Experiment Workflows
This appendix covers:
■ Advanced Setup Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 116
■ QuickStart Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 117
■ Template Workflow. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 119
■ Export/Import Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 121
Note: For more information about any of the topics discussed in this guide, access the
Help from within Applied Biosystems StepOne
™
Real-Time PCR Software v2.0 by
pressing F1, clicking in the toolbar, or selecting HelpStepOne Software Help.

Appendix A Alternate Experiment Workflows
Advanced Setup Workflow
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
116
Notes
Advanced Setup Workflow
When you create an experiment using Advanced Setup in the StepOne
™
software, you
can set up the experiment according to your own design.
1. Double-click (StepOne software shortcut) or select
StartAll ProgramsApplied BiosystemsStepOne Software
<software name>
where <software name> is the current version of the StepOne software.
2. From the Home screen, click Advanced Setup.
Note: If you do not see the Advanced Setup icon, click the arrow beneath the
Design Wizard icon to expand the Set Up menu.
3. Complete the setup screens to set up a new experiment:
a. Click Experiment Properties (default), enter the experiment name, then
select the experiment properties.
b. Click Plate Setup:
c. Click Run Method, review the reaction volume and thermal profile, then
edit as needed.
d. Click Reaction Setup, review the components and calculated volumes for
the PCR reactions, then edit as needed.
e. (Optional) Click Materials List, review the list of materials, then order
the materials you need to prepare the reaction plate.
4. Prepare the PCR reactions:
Experiment Type Action
Genotyping Define the SNP assays, then assign them to wells in the
reaction plate.
All other experiments Define the targets, then assign them to wells in the
reaction plate.
Experiment Type Action
Relative standard curve a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the standard dilution series.
d. Prepare the reaction mix.
e. Prepare the reaction plate.
Standard curve

Appendix A Alternate Experiment Workflows
QuickStart Workflow
117
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
5. Run the experiment:
a. Load the reaction plate into the instrument.
b. Start the run
c. (Optional) Monitor the run.
d. Unload the reaction plate from the instrument.
6. Analyze the data:
a. Open the experiment in the StepOne software.
b. From the Experiment Menu, click Analysis.
c. If the data are not analyzed, click Analyze.
d. In the navigation pane, select an analysis screen to view the data (for example,
select QC Summary to view a quality summary of the data).
QuickStart Workflow
When you create an experiment using QuickStart, you can run the reactions on the
instrument with no reaction plate setup information.
1. Prepare the PCR reactions:
Comparative C
T
a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the reaction mix.
d. Prepare the reaction plate.
Genotyping
Presence/absence
Experiment Type Action
Experiment Type Action
Relative standard curve a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the standard dilution series.
d. Prepare the reaction mix.
e. Prepare the reaction plate.
Standard curve
Comparative C
T
a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the reaction mix.
d. Prepare the reaction plate.
Genotyping
Presence/absence

Appendix A Alternate Experiment Workflows
QuickStart Workflow
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
118
Notes
2. QuickStart the experiment:
a. Double-click (StepOne software shortcut) or select
StartAll ProgramsApplied BiosystemsStepOne Software
<software name>
where <software name> is the current version of the StepOne software.
b. From the Home screen, click QuickStart.
c. Select the Experiment Properties tab (default), enter the experiment name,
then select the experiment properties.
d. Select the Run Method tab, review the reaction volume and thermal profile,
then edit as needed.
3. Run the experiment:
a. Load the reaction plate into the instrument.
b. Start the run
c. (Optional) Monitor the run.
d. Unload the reaction plate from the instrument.
4. In the StepOne software, complete the plate setup:
5. Analyze the data:
a. Open the experiment in the StepOne software.
b. From the Experiment Menu, click Analysis.
c. If the data are not analyzed, click Analyze.
d. In the navigation pane, select an analysis screen to view the data (for example,
select QC Summary to view a quality summary of the data).
Experiment Type Action
Genotyping a. Select and complete the Define SNP Assays and
Samples tab.
b. Select and complete the Assign SNP Assays and
Samples tab.
All other experiments a. Select and complete the Define Targets and
Samples tab.
b. Select and complete the Assign Targets and
Samples tab.

Appendix A Alternate Experiment Workflows
Templa te Wor kfl ow
119
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Template Workflow
You can use a template to create a new experiment. Templates are useful when you want
to create many experiments with the same setup information.
Create a
Template
1. Double-click (StepOne software shortcut) or select
StartAll ProgramsApplied BiosystemsStepOne Software
<software name>
where <software name> is the current version of the StepOne software.
2. Open an existing experiment, or create a new experiment.
Note: You can create a new experiment using the Design Wizard (see Chapter 2) or
Advanced Setup (see page 116).
3. Select FileSave As Template.
4. Enter a file name, select a location for the template, then click Save.
5. Click Close.
Create an
Experiment with a
Template
1. From the Home screen, click Templa te.
Note: If you do not see the Template icon, click the arrow beneath the Design
Wizard icon to expand the Set Up menu.
2. Locate and select the template you created in step d, then click Open. A new
experiment is created using the setup information from the template:
• Experiment properties
• Plate setup
• Run method
• Reaction setup
3. (Optional) If you want to modify the experiment, use Advanced Setup (see
page 116).
4. Click Save, enter a file name, then click Save to save the experiment.
5. Prepare the PCR reactions:
Experiment Type Action
Relative standard curve a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the standard dilution series.
d. Prepare the reaction mix.
e. Prepare the reaction plate.
Standard curve

Appendix A Alternate Experiment Workflows
Templa te Wor kflo w
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
120
Notes
6. Run the experiment:
a. Load the reaction plate into the instrument.
b. Start the run
c. (Optional) Monitor the run.
d. Unload the reaction plate from the instrument.
7. Analyze the data:
a. Open the experiment in the StepOne software.
b. From the Experiment Menu, click Analysis.
c. If the data are not analyzed, click Analyze.
d. In the navigation pane, select an analysis screen to view the data (for example,
select QC Summary to view a quality summary of the data).
Comparative C
T
a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the reaction mix.
d. Prepare the reaction plate.
Genotyping
Presence/absence
Experiment Type Action
Appendix A Alternate Experiment Workflows
Export/Import Workflow
121
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
Export/Import Workflow
Use the Export/Import workflow to set up a new experiment using setup data exported
from another experiment. Only reaction plate setup data are exported and imported.
Note: Setup data exported from experiments in StepOne Software v1.0 can be imported
into experiments in StepOne Software v2.0 or later. However, setup data exported from
experiments in StepOne Software v2.0 or later cannot be imported into experiments in
StepOne Software v1.0.
Export Setup
Data
1. Double-click (StepOne software shortcut) or select
StartAll ProgramsApplied BiosystemsStepOne Software
<software name>
where <software name> is the current version of the StepOne software.
2. Open an existing experiment, or create a new experiment.
Note: You can create a new experiment using the Design Wizard (see Chapter 2) or
Advanced Setup (see page 116).
3. Select FileExport.
4. Select the Export Properties tab (default), then:
a. Select Setup.
b. Select One File from the dropdown menu.
c. Enter a name, then select a location for the export file.
d. Select (*.txt) from the File Type dropdown menu.
IMPORTANT! You cannot export *.xml files.
5. (Optional) Click the Customize Export tab, then select the appropriate options.
6. Click Start Export,
7. When prompted, click Close Export Tool.
Create an
Experiment with
an Exported Text
File
You can import plate setup data from an exported text file (*.txt) to complete the reaction
plate setup data for your experiment.
IMPORTANT! Be sure the exported text file you select contains only reaction plate setup
data and that the experiment types match.

Appendix A Alternate Experiment Workflows
Export/Import Workflow
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
122
Notes
1. Import the reaction plate setup data from an exported text file:
a. Using a spreadsheet application (such as Microsoft
®
Excel software), open an
exported text file.
b. Replace the parameters of the text file as needed. When finished, save the file
as a tab-delimited text file.
c. From the Home screen, click Advanced Setup.
Note: If you do not see the Advanced Setup icon, click the arrow beneath the
Design Wizard icon to expand the Set Up menu.
d. Create a new experiment or open an existing experiment.
e. Select FileImport.
f. Click Browse, locate and select the text file (*.txt), then click Select.
g. Click Start Import. The setup data from the exported text file is imported into
the open experiment.
Note: If your experiment already contains plate setup information, the
software asks if you want to replace the plate setup with the data from the text
file. Click Ye s to replace the plate setup.
2. Use Advanced Setup to finish setting up your experiment (see page 116).
3. Prepare the PCR reactions:
4. Run the experiment:
a. Load the reaction plate into the instrument.
b. Start the run
c. (Optional) Monitor the run.
d. Unload the reaction plate from the instrument.
Experiment Type Action
Relative standard curve a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the standard dilution series.
d. Prepare the reaction mix.
e. Prepare the reaction plate.
Standard curve
Comparative C
T
a. Prepare the template.
b. Prepare the sample dilutions.
c. Prepare the reaction mix.
d. Prepare the reaction plate.
Genotyping
Presence/absence

Appendix A Alternate Experiment Workflows
Export/Import Workflow
123
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Notes
5. Analyze the data:
a. Open the experiment in the StepOne software.
b. From the Experiment Menu, click Analysis.
c. If the data are not analyzed, click Analyze.
d. In the navigation pane, select an analysis screen to view the data (for example,
select QC Summary to view a quality summary of the data).

Appendix A Alternate Experiment Workflows
Export/Import Workflow
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
124
Notes
125
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Bibliography
Kwok, S. and Higuchi, R. 1989. Avoiding false positives with PCR. Nature
339:237–238.
Saiki, R.K., Scharf, S., Faloona, F., et al. 1985. Enzymatic amplification of β-globin
genomic sequences and restriction site analysis for diagnosis of sickle cell anemia.
Science 230:1350–1354.
Bibliography
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
126
127
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Glossary
Advanced Setup In the StepOne
™
software, a feature that allows you to set up your experiment according
to your experiment design. Advanced Setup provides you with maximum flexibility in the
design and setup of your experiment.
AIF See assay information file (AIF).
allele For a given target, any of the different sequences that occurs in the population.
allelic discrimina-
tion plot
Display of data collected during the post-PCR read. The allelic discrimination plot is a
graph of the normalized reporter signal from the allele 1 probe plotted against the
normalized reporter signal from the allele 2 probe.
amplicon A segment of DNA amplified during PCR.
amplification Part of the instrument run in which PCR produces amplification of the target. For
quantitation experiments, fluorescence data collected during amplification are displayed
in an amplification plot, and the data are used to calculate results. For genotyping or
presence/absence experiments, fluorescence data collected during amplification are
displayed in an amplification plot, and the data can be used for troubleshooting.
amplification
efficiency (EFF%)
Calculation of efficiency of the PCR amplification. The amplification efficiency is
calculated using the slope of the regression line in the standard curve. A slope close to
−3.32 indicates optimal, 100% PCR amplification efficiency. Factors that affect
amplification efficiency:
• Range of standard quantities – To increase the accuracy and precision of the
efficiency measurement, use a broad range of standard quantities, 5 to 6 logs
(10
5
to 10
6
fold).
• Number of standard replicates – To increase the precision of the standard
quantities and decrease the effects of pipetting inaccuracies, include replicates.
• PCR inhibitors – PCR inhibitors in the reaction can reduce amplification and alter
measurements of the efficiency.
amplification plot Display of data collected during the cycling stage of PCR amplification. Can be viewed
as:
• Baseline-corrected normalized reporter (ΔRn) vs. cycle
• Normalized reporter (Rn) vs. cycle
• Threshold cycle (C
T
) vs. well

Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
128
amplification stage Part of the instrument run in which PCR produces amplification of the target. The
amplification stage is called a cycling stage in the thermal profile and consists of
denaturing, primer annealing, and polymerization steps that are repeated.
For quantitation experiments, fluorescence data collected during the amplification stage
are displayed in an amplification plot, and the data are used to calculate results. For
genotyping or presence/absence experiments, fluorescence data collected during the
amplification stage are displayed in an amplification plot, and the data can be used for
troubleshooting. See also cycling stage.
assay In the StepOne
™
and StepOnePlus
™
systems, a PCR reaction mix that contains primers
to amplify a target and a reagent to detect the amplified target.
Assay ID Identifier assigned by Applied Biosystems to TaqMan
®
Gene Expression Assays and
TaqMan
®
SNP Genotyping Assays.
assay information
file (AIF)
Data file on a CD shipped with each assay order. The file name includes the number from
the barcode on the plate. The information in the AIF is provided in a tab-delimited format.
assay mix PCR reaction component in Applied Biosystems TaqMan
®
Gene Expression Assays and
TaqMan
®
SNP Genotyping Assays. The assay mix contains primers designed to amplify
a target and a TaqMan
®
probe designed to detect amplification of the target.
AutoDelta In the run method, a setting to increase or decrease the temperature and/or time for a step
with each subsequent cycle in a cycling stage. When AutoDelta is enabled for a cycling
stage, the settings are indicated by an icon in the thermal profile:
• AutoDelta on:
• AutoDelta off:
automatic baseline An analysis setting in which the software calculates the baseline start and end values for
the amplification plot. You can apply the automatic baseline setting to specific wells in
the reaction plate. See also baseline.
automatic C
T
An analysis setting in which the software calculates the baseline start and end values and
the threshold in the amplification plot. The software uses the baseline and threshold to
calculate the threshold cycle (C
T
). See also threshold cycle (C
T
).
baseline In the amplification plot, a line fit to the fluorescence levels during the initial stages of
PCR, when there is little change in fluorescence signal.

Glossary
129
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
baseline-corrected
normalized reporter
(
ΔRn)
The magnitude of normalized fluorescence signal generated by the reporter:
1. In experiments that contain data from real-time PCR, the magnitude of normalized
fluorescence signal generated by the reporter at each cycle during the PCR
amplification. In the
ΔRn vs. Cycle amplification plot, ΔRn is calculated at each
cycle as:
ΔRn (cycle) = Rn (cycle) − Rn (baseline), where Rn = normalized reporter
2. In genotyping experiments and presence/absence experiments, the difference in
normalized fluorescence signal generated by the reporter between the pre-PCR read
and the post-PCR read. In the allelic discrimination plot (genotyping experiments)
and the presence/absence plot (presence/absence experiments), ΔRn is calculated
as:
ΔRn = Rn (post-PCR read) − Rn (pre-PCR read), where Rn = normalized reporter
See also normalized reporter (Rn).
blocked IPC In presence/absence experiments, a reaction that contains IPC blocking agent, which
blocks amplification of the internal positive control (IPC). In the StepOne
™
software, the
task for the IPC target in wells that contain IPC blocking agent. See also negative control-
blocked IPC wells.
calibrator See reference sample.
chemistry See reagents.
colocated layout A system layout in which the StepOne
™
or StepOnePlus
™
instrument is directly
connected to a colocated computer by the yellow cable. In this layout, you can control the
instrument with the StepOne
™
software on the colocated computer or with the instrument
touchscreen.
comparative C
T
(
ΔΔC
T
) method
Method for determining relative target quantity in samples. With the comparative C
T
(
ΔΔC
T
) method, the StepOne
™
software measures amplification of the target and of the
endogenous control in samples and in a reference sample. Measurements are normalized
using the endogenous control. The software determines the relative quantity of target in
each sample by comparing normalized target quantity in each sample to normalized target
quantity in the reference sample.
C
T
See threshold cycle (C
T
).
custom dye Dye that is not supplied by Applied Biosystems. Custom dyes may be adapted for use in
experiments on the StepOne
™
and StepOnePlus
™
systems. When using custom dyes, the
custom dye should be added to the Dye Library and a custom dye calibration performed.
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA
™
dye as
reporter or quencher with the StepOne
™
system. TAMRA dye may be used as a reporter
or quencher with the StepOnePlus
™
system.
cycle threshold See threshold cycle (C
T
).

Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
130
cycling stage In the thermal profile, a stage that is repeated. A cycling stage is also called an
amplification stage. For cycling stages, you can enable AutoDelta settings. See also
amplification stage.
data collection A process during the instrument run in which an instrument component detects
fluorescence data from each well of the reaction plate. The instrument transforms the
signal to electronic data, and the data are saved in the experiment file. In the StepOne
™
software, a data collection point is indicated by an icon in the thermal profile:
• Data collection on:
• Data collection off:
delta Rn (
ΔRn) See baseline-corrected normalized reporter (ΔRn).
derivative reporter
(−Rn′)
The negative first-derivative of the normalized fluorescence generated by the reporter
during PCR amplification. In the derivative reporter (–Rn′) vs. temperature melt curve,
the derivative reporter signal is displayed in the y-axis.
Design Wizard A feature in the StepOne
™
software that helps you set up your experiment by guiding you
through best practices as you enter your experiment design.
diluent A reagent used to dilute a sample or standard before adding it to the PCR reaction. The
diluent can be water or buffer.
Diluted Sample
Concentration (10✕
for Reaction Mix)
In the StepOne
™
software, a field displayed on the Sample Dilution Calculations tab of
the Reaction Setup screen. For this field, enter the sample concentration you want to use
to add to the reaction mix for all samples in the experiment. “10✕ for Reaction Mix”
indicates that the software assumes the sample or standard component of the reaction mix
is at a 10✕ concentration. For example, if the diluted sample concentration is 50.0 ng/μL
(10✕), the final sample concentration in the reaction is 5 ng/μL (1✕).
dilution factor See serial factor.
dissociation curve See melt curve.
EFF% See amplification efficiency (EFF%).
endogenous
control
A target or gene that should be expressed at similar levels in all samples you are testing.
Endogenous controls are used in relative standard curve and comparative C
T
(ΔΔC
T
)
experiments to normalize fluorescence signals for the target you are quantifying.
Housekeeping genes can be used as endogenous controls. See also housekeeping gene.
endpoint read See post-PCR read.
Glossary
131
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
experiment Refers to the entire process of performing a run using the StepOne
™
or StepOnePlus
™
systems, including setup, run, and analysis. The types of experiments you can perform
using the StepOne and StepOnePlus systems:
• Quantitation - standard curve
• Quantitation - relative standard curve
• Quantitation - comparative C
T
(ΔΔC
T
)
•Melt curve
•Genotyping
• Presence/absence
experiment name Entered during experiment setup, the name that is used to identify the experiment.
Experiment names cannot exceed 100 characters and cannot include any of the following
characters: forward slash (/), backslash (\), greater than sign (>), less than sign (<),
asterisk (*), question mark (?), quotation mark ("), vertical line (|), colon (:), or
semicolon (;).
experiment type The type of experiment you are performing using the StepOne
™
or StepOnePlus
™
system:
• Standard curve
• Comparative C
T
(ΔΔC
T
)
• Relative standard curve
• Melt curve (not available in the Design Wizard)
•Genotyping
• Presence/absence
The experiment type you select affects the setup, run, and analysis.
forward primer Oligonucleotide that flanks the 5′ end of the amplicon. The reverse primer and the forward
primer are used together in PCR reactions to amplify the target.
holding stage In the thermal profile, a stage that includes one or more steps. You can add a holding stage
to the thermal profile to activate enzymes, to inactivate enzymes, or to incubate a reaction.
housekeeping gene A gene that is involved in basic cellular functions and is constitutively expressed.
Housekeeping genes can be used as endogenous controls. See also endogenous control.
internal positive
control (IPC)
In presence/absence experiments, a short synthetic DNA template that is added to PCR
reactions. You can use the IPC to distinguish between true negative results (that is, the
target is absent in the samples) and negative results caused by PCR inhibitors, incorrect
assay setup, or reagent or instrument failure.
inventoried assays TaqMan
®
Gene Expression Assays and TaqMan
®
SNP Genotyping Assays that have been
previously manufactured, passed quality control specifications, and stored in inventory.
IPC In presence/absence experiments, abbreviation for internal positive control (IPC). In the
StepOne
™
software, the task for the IPC target in wells that contain the IPC and do not
contain IPC blocking agent. See also internal positive control (IPC).
IPC blocking agent Reagent added to PCR reactions to block amplification of the internal positive control
(IPC).
Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
132
IPC+ See negative control-IPC wells.
made-to-order
assays
TaqMan
®
Gene Expression Assays or TaqMan
®
SNP Genotyping Assays that are
manufactured at the time of order. Only assays that pass manufacturing quality control
specifications are shipped.
manual baseline An analysis setting in which you enter the baseline start and end values for the
amplification plot. You can apply the manual baseline setting to specific wells in the
reaction plate.
manual C
T
An analysis setting in which you enter the threshold value and select whether to use
automatic baseline or manual baseline values. The software uses the baseline and the
threshold values to calculate the threshold cycle (C
T
).
melt curve A plot of data collected during the melt curve stage. Peaks in the melt curve can indicate
the melting temperature (Tm) of the target or can identify nonspecific PCR amplification.
In the StepOne
™
software, you can view the melt curve as normalized reporter (Rn) vs.
temperature or as derivative reporter (−Rn′) vs. temperature. Also called dissociation
curve.
melt curve stage In the thermal profile, a stage with a temperature increment to generate a melt curve.
melting
temperature (Tm)
In melt curve experiments, the temperature at which 50% of the DNA is double-stranded
and 50% of the DNA is dissociated into single-stranded DNA. The Tm is displayed in the
melt curve.
multicomponent
plot
A plot of the complete spectral contribution of each dye for the selected well(s) over the
duration of the PCR run.
negative control
(NC)
In the StepOne
™
software, the task for targets or SNP assays in wells that contain water
or buffer instead of sample. No amplification of the target should occur in negative control
wells. Previously called no template control (NTC).
negative control-
blocked IPC wells
In presence/absence experiments, wells that contain IPC blocking agent instead of sample
in the PCR reaction. No amplification should occur in negative control-blocked IPC wells
because the reaction contains no sample and amplification of the IPC is blocked.
Previously called no amplification control (NAC).
negative control-
IPC wells
In presence/absence experiments, wells that contain IPC template and buffer or water
instead of sample. Only the IPC template should amplify in negative control-IPC wells
because the reaction contains no sample. Previously called IPC+.
no amplification
control (NAC)
See negative control-blocked IPC wells.
no template control
(NTC)
See negative control (NC).
Glossary
133
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
nonfluorescent
quencher-minor
groove binder
(NFQ-MGB)
Molecules that are attached to the 3′ end of TaqMan
®
probes. When the probe is intact,
the nonfluorescent quencher (NFQ) prevents the reporter dye from emitting fluorescence
signal. Because the NFQ does not fluoresce, it produces lower background signals,
resulting in improved precision in quantitation. The minor groove binder (MGB)
increases the melting temperature (Tm) without increasing probe length. It also allows the
design of shorter probes.
normalized quantity Quantity of target divided by the quantity of endogenous control.
normalized reporter
(Rn)
Fluorescence signal from the reporter dye normalized to the fluorescence signal of the
passive reference.
omit well An action that you perform before reanalysis to omit one or more wells from analysis.
Because no algorithms are applied to omitted wells, omitted wells contain no results.
outlier For a set of data, a datapoint that is significantly smaller or larger than the others.
passive reference A dye that produces fluorescence signal. Because the passive reference signal should be
consistent across all wells, it is used to normalize the reporter dye signal to account for
non-PCR related fluorescence fluctuations caused by minor well-to-well differences in
concentrations or volume. Normalization to the passive reference signal allows for high
data precision.
plate layout An illustration of the grid of wells and assigned content in the reaction plate. In StepOne
™
systems, the grid contains 6 rows and 8 columns. In StepOnePlus
™
systems, the grid
contains 8 rows and 12 columns.
In the StepOne
™
software, you can use the plate layout as a selection tool to assign well
contents, to view well assignments, and to view results. The plate layout can be printed,
included in a report, exported, and saved as a slide for a presentation.
point One standard in a standard curve. The standard quantity for each point in the standard
curve is calculated based on the starting quantity and serial factor.
positive control In genotyping experiments, a DNA sample with a known genotype, homozygous or
heterozygous. In the StepOne
™
software, the task for the SNP assay in wells that contain
a sample with a known genotype.
post-PCR read Used in genotyping and presence/absence experiments, the part of the instrument run that
occurs after amplification. In genotyping experiments, fluorescence data collected during
the post-PCR read are displayed in the allelic discrimination plot and used to make allele
calls. In presence/absence experiments, fluorescence data collected during the post-PCR
read are displayed in the presence/absence plot and used to make detection calls. Also
called endpoint read.
pre-PCR read Used in genotyping and presence/absence experiments, the part of the instrument run that
occurs before amplification. The pre-PCR read is optional but recommended.
Fluorescence data collected during the pre-PCR read can be used to normalize
fluorescence data collected during the post-PCR read.
primer mix PCR reaction component that contains the forward primer and reverse primer designed to
amplify the target.

Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
134
primer/probe mix PCR reaction component that contains the primers designed to amplify the target and a
TaqMan
®
probe designed to detect amplification of the target.
pure dye See custom dye and system dye.
quantitation
method
In quantitation experiments, the method used to determine the quantity of target in the
samples. In StepOne
™
and StepOnePlus
™
systems, there are three types of quantitation
methods: standard curve, relative standard curve, and comparative C
T
(ΔΔC
T
).
quantity In quantitation experiments, the amount of target in the samples. Absolute quantity can
refer to copy number, mass, molarity, or viral load. Relative quantity refers to the fold-
difference between normalized quantity of target in the sample and normalized quantity
of target in the reference sample.
quencher A molecule attached to the 3′ end of TaqMan
®
probes to prevent the reporter from
emitting fluorescence signal while the probe is intact. With TaqMan
®
reagents, a
nonfluorescent quencher-minor groove binder (NFQ-MGB) can be used as the quencher.
With SYBR
®
Green reagents, no quencher is used.
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA
™
dye as
reporter or quencher with the StepOne
™
system. TAMRA dye may be used as a reporter
or quencher with the StepOnePlus
™
system.
QuickStart A feature in StepOne
™
and StepOnePlus
™
systems that allows you to run an experiment
without entering plate setup information. QuickStart requires a colocated layout with the
instrument powered on and an intact instrument-computer connection.
R
2
value Regression coefficient calculated from the regression line in the standard curve. The R
2
value indicates the closeness of fit between the standard curve regression line and the
individual C
T
data points from the standard reactions. A value of 1.00 indicates a perfect
fit between the regression line and the data points.
ramp The rate at which the temperature changes during the instrument run. Except for the melt
curve step, the ramp is defined as a percentage. For the melt curve step, the ramp is
defined as a temperature increment. In the graphical view of the thermal profile, the ramp
is indicated by a diagonal line.
ramp speed Speed at which the temperature ramp occurs during the instrument run. Available ramp
speeds include fast and standard.
• For optimal results using the fast ramp speed, Applied Biosystems recommends
using TaqMan
®
Fast reagents in your PCR reactions.
• For optimal results using the standard ramp speed, Applied Biosystems
recommends using standard reagents in your PCR reactions.
IMPORTANT! TaqMan Fast reagents are not supported for genotyping or
presence/absence experiments.
raw data plot A plot of raw fluorescence signal (not normalized) for each optical filter.
Glossary
135
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
reaction mix A solution that contains all components to run the PCR reaction, except for the template
(sample, standard, or control).
reagents The PCR reaction components you are using to amplify the target and to detect
amplification. Types of reagents used on the StepOne
™
and StepOnePlus
™
systems:
•TaqMan
®
reagents
• SYBR
®
Green reagents
• Other reagents
real-time PCR Process of collecting fluorescence data during PCR. Data from the real-time PCR are used
to calculate results for quantitation experiments or to troubleshoot results for genotyping
or presence/absence experiments.
reference sample In relative standard curve and comparative C
T
(ΔΔC
T
) experiments, the sample used as the
basis for relative quantitation results. Also called the calibrator.
refSNP ID Identifies the reference SNP (refSNP) cluster ID. Generated by the Single Nucleotide
Polymorphism Database of Nucleotide Sequence Variation (dbSNP) at the National
Center for Biotechnology Information (NCBI). The refSNP ID can be used to search the
Applied Biosystems Store for an Applied Biosystems SNP Genotyping Assay. Also called
an rs number.
regression
coefficients
Values calculated from the regression line in standard curves, including the R
2
value,
slope, and y-intercept. You can use the regression coefficients to evaluate the quality of
results from the standards. See also standard curve.
regression line In standard curve and relative standard curve experiments, the best-fit line from the
standard curve. Regression line formula:
C
T
= m [log (Qty)] + b
where m is the slope, b is the y-intercept, and Qty is the standard quantity.
See also regression coefficients.
reject well An action that the software performs during analysis to remove one or more wells from
further analysis if a specific flag is applied to the well. Rejected wells contain results
calculated up to the point of rejection.
relative standard
curve method
Method for determining relative target quantity in samples. With the relative standard
curve method, the StepOne
™
software measures amplification of the target and of the
endogenous control in samples, in a reference sample, and in a standard dilution series.
Measurements are normalized using the endogenous control. Data from the standard
dilution series are used to generate the standard curve. Using the standard curve, the
software interpolates target quantity in the samples and in the reference sample. The
software determines the relative quantity of target in each sample by comparing target
quantity in each sample to target quantity in the reference sample.
Remote Monitor A feature in the StepOne
™
software that allows you to monitor a StepOne
™
or
StepOnePlus
™
instrument over the network. With the Remote Monitor, you can monitor
the instrument status, send an experiment to the instrument, monitor amplification plots
and temperature plots in real time, and download the results to your computer. You cannot
operate the StepOne or StepOnePlus instrument using the Remote Monitor.
Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
136
replicate group A set of identical reactions in an experiment.
replicates Total number of identical reactions containing identical components and identical
volumes.
reporter Fluorescent dye used to detect amplification. If you are using TaqMan
®
reagents, the
reporter dye is attached to the 5′ end. If you are using SYBR
®
Green reagents, the reporter
dye is SYBR
®
Green dye.
reverse primer An oligonucleotide that flanks the 3′ end of the amplicon. The reverse primer and the
forward primer are used together in PCR reactions to amplify the target.
reverse
transcriptase
An enzyme that converts RNA to cDNA. Reverse transcriptase is added to the PCR
reaction to perform 1-step RT-PCR.
Rn See normalized reporter (Rn).
ROX
™
dye A dye supplied by Applied Biosystems and precalibrated on the StepOne
™
and
StepOnePlus
™
systems. ROX dye is used as the passive reference.
rs number See refSNP ID.
run method Definition of the reaction volume and the thermal profile for the StepOne
™
or
StepOnePlus
™
instrument run.
sample The template that you are testing.
Sample DNA (10✕) In the StepOne™ software, a reaction component displayed on the Reaction Mix
Calculations tab of the Reaction Setup screen. The software assumes the sample DNA is
added to the reaction mix at a 10✕ concentration. For example, if the reaction volume is
20 μL, the calculated volume of sample for 1 reaction is 2 μL.
Sample Library In the StepOne
™
software, a collection of samples. The Sample Library contains the
sample name and the sample color.
Sample or Standard
(10✕ )
In the StepOne
™
software, a reaction component displayed on the Reaction Mix
Calculations tab of the Reaction Setup screen. The software assumes the sample or
standard is added to the reaction mix at a 10✕ concentration. For example, if the reaction
volume is 20 μL, the calculated volume of sample or standard for 1 reaction is 2 μL.
sample/SNP assay
reaction
In genotyping experiments, the combination of which sample to test and which SNP assay
to perform in one PCR reaction. Each PCR reaction can contain only one sample and one
SNP assay.
sample/target
reaction
In quantitation experiments, the combination of which sample to test and which target to
detect and quantify in one PCR reaction. In the Design Wizard, you can detect and
quantify only one target in one PCR reaction. Use Advanced Setup to detect and quantify
more than one target in one PCR reaction.
Glossary
137
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
serial factor In the StepOne
™
software, a numerical value that defines the sequence of quantities in the
standard curve. The serial factor and the starting quantity are used to calculate the
standard quantity for each point in the standard curve. For example, if the standard curve
is defined with a serial factor of 1:10 or 10✕, the difference between any 2 adjacent points
in the curve is 10-fold.
series See standard dilution series.
slope Regression coefficient calculated from the regression line in the standard curve. The slope
indicates the PCR amplification efficiency for the assay. A slope of −3.32 indicates 100%
amplification efficiency. See also amplification efficiency (EFF%) and regression line.
SNP Abbreviation for single nucleotide polymorphism. The SNP can consist of a base
difference or an insertion or deletion of one base.
SNP assay Used in genotyping experiments, a PCR reaction that contains primers to amplify the SNP
and two probes to detect different alleles.
SNP Assay Library In the StepOne
™
software, a collection of SNP assays to add to genotyping experiments.
The SNP assays in the library contain the SNP assay name, SNP assay color, and for each
allele, the allele name or base(s), reporter, quencher, and allele colors. The SNP assays in
the library may also contain the assay ID and comments about the SNP assay.
spatial calibration Ty p e o f StepOne
™
and StepOnePlus
™
system calibration in which the system maps the
positions of the wells in the sample block(s). Spatial calibration data are used so that the
software can associate increases in fluorescence during a run with specific wells in the
reaction plate.
stage In the thermal profile, a group of one or more steps. There are three types of stages:
holding stage (including pre-PCR read and post-PCR read), cycling stage (also called
amplification stage), and melt curve stage.
standalone layout A system layout in which the StepOne
™
or StepOnePlus
™
instrument is not connected to
a computer by the yellow cable. In this layout, you control the instrument only with the
instrument touchscreen, and you use a USB drive or network connection to transfer data
between the instrument and computer.
standard Sample that contains known standard quantities. Standard reactions are used in
quantitation experiments to generate standard curves. See also standard curve and
standard dilution series.
standard curve In standard curve and relative standard curve experiments:
• The best-fit line in a plot of the C
T
values from the standard reactions plotted
against standard quantities. See also regression line.
• A set of standards containing a range of known quantities. Results from the standard
curve reactions are used to generate the standard curve. The standard curve is
defined by the number of points in the dilution series, the number of standard
replicates, the starting quantity, and the serial factor. See also standard dilution
series.
Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
138
standard curve
method
Method for determining absolute target quantity in samples. With the standard curve
method, the StepOne
™
software measures amplification of the target in samples and in a
standard dilution series. Data from the standard dilution series are used to generate the
standard curve. Using the standard curve, the software interpolates the absolute quantity
of target in the samples. See also standard and standard curve.
standard dilution
series
In standard curve and relative standard curve experiments, a set of standards containing a
range of known quantities. The standard dilution series is prepared by serially diluting
standards. For example, the standard stock is used to prepare the first dilution point, the
first dilution point is used to prepare the second dilution point, and so on. In the StepOne
™
software, the volumes needed to prepare a standard dilution series are calculated by the
number of dilution points, the number of standard replicates, the starting quantity, the
serial factor, and the standard concentration in the stock. See also standard curve.
standard quantity A known quantity in the PCR reaction.
• In standard curve experiments, the quantity of target in the standard. In the
StepOne
™
software, the units for standard quantity can be for mass, copy number,
viral load, or other units for measuring the quantity of target.
• In relative standard curve experiments, a known quantity in the standard. Standard
quantity can refer to the quantity of cDNA or the quantity of standard stock in the
PCR reaction. The units are not relevant for relative standard curve experiments
because they cancel out in the calculations.
starting quantity When defining a standard curve in the StepOne
™
software, corresponds to the highest or
lowest quantity.
step A component of the thermal profile. For each step in the thermal profile, you can set the
ramp rate (ramp increment for melt curve steps), hold temperature, hold time (duration),
and you can turn data collection on or off for the ramp or the hold parts of the step. For
cycling stages, a step is also defined by the AutoDelta status. With StepOnePlus
™
systems, which contain the VeriFlex
™
blocks, each step contains 6 temperatures (1 for
each VeriFlex block).
SYBR
®
Green
reagents
PCR reaction components that consist of two primers designed to amplify the target and
SYBR
®
Green dye to detect double-stranded DNA.

Glossary
139
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
system dye Dye supplied by Applied Biosystems and precalibrated on the StepOne
™
or
StepOnePlus
™
system. Before you use system dyes in your experiments, make sure the
system dye calibration is current in the Instrument Maintenance Manager.
System dyes on the StepOne system:
•FAM
™
dye
•JOE
™
dye
•ROX
™
dye
• SYBR
®
Green dye
•VIC
®
dye
System dyes on the StepOnePlus system:
•FAM
™
dye
•JOE
™
dye
•NED
™
dye
•ROX
™
dye
• SYBR
®
Green dye
• TAMRA
™
dye
•VIC
®
dye
IMPORTANT! Applied Biosystems does not recommend the use of TAMRA
™
dye as
reporter or quencher with the StepOne
™
system. TAMRA dye may be used as a reporter
or quencher with the StepOnePlus
™
system.
TaqMan
®
reagents PCR reaction components that consist of primers designed to amplify the target and a
TaqMan
®
probe designed to detect amplification of the target.
target The nucleic acid sequence that you want to amplify and detect.
target color In the StepOne
™
software, a color assigned to a target to identify the target in the plate
layout and analysis plots.
Target Library In the StepOne
™
software, a collection of targets to add to experiments. The targets in the
library contain the target name, reporter, quencher, and target color. The target in the
library may also contain comments about the target.
task In the StepOne
™
software, the type of reaction performed in the well for the target or SNP
assay. Available tasks:
• Unknown
•Negative Control
• Standard (standard curve and relative standard curve experiments)
• Positive control (genotyping experiments)
• IPC (presence/absence experiments)
• Blocked IPC (presence/absence experiments)
Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
140
temperature plot In the StepOne
™
software, a display of temperatures for the sample, instrument cover, and
instrument block during the StepOne
™
or StepOnePlus
™
instrument run.
template In the Design Wizard of the StepOne
™
software (and in QuickStart for quantitation
experiments), the type of nucleic acid to add to the PCR reaction. The recommended
template varies according to experiment type:
• Quantitation experiments (standard curve, relative standard curve, and comparative
C
T
) – cDNA (complementary cDNA), RNA, or gDNA (genomic DNA)
For quantitation experiments, the template type selection affects the run method,
reaction setup, and materials list.
• Genotyping experiments – Wet DNA (gDNA or cDNA) or dry DNA (gDNA or
cDNA)
For genotyping experiments, the template type selection affects the reaction setup.
• Presence/absence experiments - DNA
For presence/absence experiments, Applied Biosystems recommends adding DNA
templates to the PCR reactions.
thermal profile Part of the run method that specifies the temperature, time, ramp, and data collection
points for all steps and stages of the StepOne
™
or StepOnePlus
™
instrument run.
threshold 1. In amplification plots, the level of fluorescence above the baseline and within the
exponential growth region The threshold can be determined automatically (see
automatic C
T
) or can be set manually (see manual C
T
).
2. In presence/absence experiments, the level of fluorescence above which the
StepOne
™
software assigns a presence call.
threshold cycle (C
T
) The PCR cycle number at which the fluorescence meets the threshold in the amplification
plot.
Tm See melting temperature (Tm).
touchscreen Instrument display that you touch to control the StepOne
™
or StepOnePlus
™
instrument.
unknown In the StepOne
™
software, the task for the target or SNP assay in wells that contain the
sample you are testing:
• In quantitation experiments, the task for the target in wells that contain a sample
with unknown target quantities.
• In genotyping experiments, the task for the SNP assay in wells that contain a sample
with an unknown genotype.
• In presence/absence experiments, the task for the target in wells that contain a
sample in which the presence of the target is not known.
unknown-IPC wells In presence/absence experiments, wells that contain a sample and internal positive control
(IPC).

Glossary
141
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
VeriFlex
™
Technology
The StepOnePlus
™
instrument contains six independently thermally regulated VeriFlex
™
blocks, creating up to six different zones for the 96 sample wells. After you enable the
VeriFlex blocks in the StepOne
™
software, you can set a different temperature for one or
more of the VeriFlex blocks.
y-intercept In the standard curve, the value of y where the regression line crosses the y-axis. The y-
intercept indicates the expected threshold cycle (C
T
) for a sample with quantity equal to 1.
zone One of up to six sample temperatures among the 96 wells formed by independently
thermally regulated VeriFlex
™
blocks during the StepOnePlus
™
instrument run. You can
set a different temperature for one or more of the VeriFlex blocks, or you can set the same
temperature for each of the VeriFlex blocks.
Note: For melt curve steps, you need to set the same temperature for each of the
VeriFlex blocks.
zone boundary The edge of a zone for samples formed by the six independently thermally regulated
Ver iF lex
™
blocks. In the StepOne
™
software, the zone boundaries are displayed in the
plate layout as thick red lines.
Glossary
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
142
143
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
Index
Numerics
1-step RT-PCR 7, 24, 32, 37
2-step RT-PCR 7
, 11, 24
A
Advanced Setup 9, 10, 24, 30, 56, 116
alternate experiment workflows. See workflows.
amplification efficiency 28
, 92
Amplification Plot screen
monitor during a run 70
view after a run 92
amplification plot, typical 96
AMPNC flag 107
analysis screens
Amplification Plot screen 92
Multicomponent Plot screen 109
Multiple Plots screen 89
navigation tips 88
QC Summary screen 106
Raw Data Plot screen 112
software elements 85
Standard Curve screen 90
Well Table 99
analysis settings
advanced 105
baseline 105
CT 105
flag 105
threshold 105
view 104
analyze experiment
analyze 84
for more information 92
, 98, 101, 105, 107, 109,
111, 113
guidelines 85
, 92, 96, 101, 105, 107, 109, 111, 113
omit wells 108
publish the data 102
view Amplification Plot screen 92
view analysis settings 104
view Multicomponent Plot screen 109
view Multiple Plots screen 89
view QC Summary screen 106
view Raw Data Plot screen 112
view Standard Curve screen 90
view Well Table 99
workflow 82
Applied Biosystems
contacting xii
customer feedback on documentation xii
Technical Support xii
Applied Biosystems StepOne Real-Time PCR System.
See StepOne system.
assumptions for using this guide viii
B
BADROX flag 107
baseline
correct values 96
examples 98
manually adjust 105
biohazardous waste, handling xx
BLFAIL flag 107
C
CAUTION, description xiii
chemical safety xviii
chemical waste safety xix
colocated layout
data transfer 77
monitoring 69
startup 65
consumables
Also see materials required 4
supported 4
conventions used in this guide viii
create new experiment 17
CT 101
CTFAIL flag 107
Custom assays 37
customer feedback, on Applied Biosystems
documents xii
D
DANGER, description xiii
data
about data collection 2
example experiment 9
, 84
publish 102
transfer 76
design experiment
create new 17
define experiment properties 20
define methods and materials 22
finish Design Wizard 42
for more information 22
, 24, 26, 28, 31, 33, 38, 41,
Index
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
144
44
guidelines 21
, 23, 26, 28, 30, 32, 37, 41, 44
order materials 38
review reaction setup 33
set up run method 31
set up samples 29
set up standards 27
set up targets 25
software elements 18
workflow 16
Design Wizard
Experiment Properties screen 20
finish 42
Materials List screen 38
Methods & Materials screen 22
Reaction Setup screen 33
Run Method screen 31
Samples screen 29
software elements 18
Standards screen 27
Targets screen 25
deviation, standard 97
documentation, related ix
E
electrical safety xx
electromagnetic compatibility standards. See EMC
standards
EMC standards xxii
ergonomics, safety xxii
example experiment
analyze 82
data 12
data location 12
description 10
design 16
name 20
prepare 46
run 60
workflow 13
Experiment Properties screen 20
EXPFAIL flag 107
Export/Import 10
, 121
F
flag 101
flags
analysis settings 105
in standard curve experiments 107
G
General xxi
guidelines
analysis 85
, 92, 96, 101, 105, 107, 109, 111, 113
chemical safety xviii
chemical waste disposal xix
chemical waste safety xix
design 21
, 23, 26, 28, 30, 32, 37, 41, 44
preparation 48
, 50, 52, 56
run 64
H
hazard icons. See safety symbols, on instruments
hazard symbols. See safety symbols, on instruments
hazards. See safety
Help system, accessing xi
HIGHSD flag 107
I
installation category xxi
instrument operation, safety xvii
Inventoried assays 37
L
library 32
load reaction plate 62
M
Made to Order assays 37
Materials List screen 38
materials required 47
, 49, 51, 54
Methods & Materials screen 22
monitor run
Amplification Plot screen 70
colocated layout 69
Run Method screen 72
standalone layout 75
standalone layout (remote) 73
Temperature Plot screen 71
moving and lifting, safety xvii
MSDSs
description xviii
obtaining xviii
MSDSs, obtaining xii
MTP flag 107
Multicomponent Plot screen 109
Multiple Plots screen 89
multiplex PCR 6
, 24
N
navigation tips
display multiple plots 89
select wells 88
negative control, component of experiment 6
NOAMP flag 107
NOISE flag 107
Index
145
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
NOSIGNAL flag 107
notification settings 63
O
OFFSCALE flag 107
omit wells 108
online Help. See Help system
order materials 38
other fluorescent-based reagents 9
OUTLIERRG flag 107
outliers. See omit wells.
overvoltage category (rating) xxi
P
prepare experiment
for more information 48
, 53, 57
guidelines 48
, 50, 52, 56
reaction mix
prepare 51
reaction plate 53
sample dilutions 47
standard dilution series 49
workflow 46
prepare for run 61
print reaction setup instructions 36
publish data 102
Q
QC Summary screen 106
QuickStart 10
, 117
R
radioactive waste, handling xx
ramp speed 22
, 24
Raw Data Plot screen 112
reaction mix
calculated volumes 34
volumes 51
reaction plate
layout 42
load 62
prepare 53
unload from the instrument 76
Reaction Setup screen 33
reagents
other fluorescent-based 9
SYBR Green 8
TaqMan 8
repetitive motion, safety xxii
replicate, component of experiment 6
results, interpreting 83
run experiment
alerts 72
enable notification settings 63
for more information 63
, 72, 73
guidelines 64
monitor 69
prepare for 61
start 65
transfer data 76
workflow 60
Run Method library 32
Run Method screen 31
monitor during a run 72
S
safety
before operating the instrument xvii
biological hazards xxi
chemical xviii
chemical waste xix
conventions xiii
electrical xx
ergonomic xxii
guidelines xviii
, xix, xx
instrument operation xvii
moving and lifting instrument xvii
moving/lifting xvii
repetitive motion xxii
standards xxii
workstation xxii
safety labels, on instruments xvi
safety standards xxii
safety symbols, on instruments xv
sample dilutions
calculated volumes 36
, 47
prepare 47
Sample screen 29
samples
design guidelines 30
dilutions 47
sample reactions (unknowns) 55
set up 29
select wells 88
singleplex PCR 6
, 24
software elements
analysis screens 85
Design Wizard 18
SPIKE flag 107
standalone layout
data transfer 78
monitoring 75
remote data transfer 77
Remote Monitor 73
startup 66
standard curve experiments
about 6
standard deviation, effect of threshold on 97
Index
Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Getting Started Guide for Standard
Curve Experiments
146
standard dilution series
calculated volumes 35
component of experiment 6
prepare 49
standards
component of experiment 6
design guidelines 28
diluting 49
EMC xxii
safety xxii
set up 27
Set Up Standards checkbox 25
, 26
standard reactions 55
Standards screen 27
start run
colocated layout 65
standalone layout 66
StepOne system
consumables 4
data collection 2
filters 3
, 113
layouts 65
, 69, 76
reagents 8
SYBR Green reagents 3
, 8, 24, 26, 37, 113
symbols, safety xv
T
TaqMan reagents 3, 8, 23, 24, 26, 37, 113
targets
design guidelines 26
set up 25
Targets screen 25
Technical Support, contacting xii
Temperature Plot screen 71
Template 10
, 119
template. See samples.
text conventions viii
THOLDFAIL flag 107
threshold
correct values 96
examples 97
manually adjust 105
training, information on xii
transfer data 76
troubleshooting
adjust baseline 105
adjust threshold 105
flags 107
omit wells 108
view analysis settings 104
view Multicomponent Plot screen 109
view QC Summary screen 106
view Raw Data Plot screen 112
U
unload instrument 76
user attention words, described viii
using this guide
as a tutorial 9
with your own experiments 9
W
WARNING, description xiii
waste disposal, guidelines xx
waste profiles, description xx
Well Table 99
wells
negative control 29
, 42, 53
omit 108
selecting 88
standard 29
, 42, 53
unknown 29
, 42, 53
workflows
Advanced Setup 116
example experiment 13
Export/Import 10
, 121
QuickStart 10
, 117
Template 10
, 119
workstation safety xxii

www.appliedbiosystems.com
Worldwide Sales and Support
Applied Biosystems vast distribution and
service network, composed of highly trained
support and applications personnel,
reaches 150 countries on six continents.
For sales office locations and technical support,
please call our local office or refer to our
Web site at www.appliedbiosystems.com.
Headquarters
850 Lincoln Centre Drive
Foster City, CA 94404 USA
Phone: +1 650.638.5800
Tol l F re e (I n No rth Am er ica ) : +1 8 00 .3 45 .52 2 4
Fax: +1 650.638.5884
08/2010
Part Number 4376784 Rev. F


