
Predicting microbial growth conditions from amino acid composition
Tyler P. Barnum
1
, Alexander Crits-Christoph
1
, Michael Molla
1
, Paul Carini
1,2
, Henry H. Lee
1
, Nili
Ostrov
1
1
Cultivarium, 490 Arsenal Way, Watertown MA 02472, USA
2
Department of Environmental Science, University of Arizona, Tucson AZ 85750
Abstract
The ability to grow a microbe in the laboratory enables reproducible study and engineering of its
genetics. Unfortunately, the majority of microbes in the tree of life remain uncultivated because of
the effort required to identify culturing conditions. Predictions of viable growth conditions to guide
experimental testing would be highly desirable. While carbon and energy sources can be
computationally predicted with annotated genes, it is harder to predict other requirements for
growth such as oxygen, temperature, salinity, and pH. Here, we developed genome-based
computational models capable of predicting oxygen tolerance (92% balanced accuracy), optimum
temperature (R
2
=0.73), salinity (R
2
=0.81) and pH (R
2
=0.48) for novel taxonomic microbial families
without requiring functional gene annotations. Using growth conditions and genome sequences of
15,596 bacteria and archaea, we found that amino acid frequencies are predictive of growth
requirements. As little as two amino acids can predict oxygen tolerance with 88% balanced
accuracy. Using cellular localization of proteins to compute amino acid frequencies improved
prediction of pH (R
2
increase of 0.36). Because these models do not rely on the presence or
absence of specific genes, they can be applied to incomplete genomes, requiring as little as 10%
completeness. We applied our models to predict growth requirements for all 85,205 species of
sequenced bacteria and archaea and found that uncultivated species are enriched in thermophiles,
anaerobes, and acidophiles. Finally, we applied our models to 3,349 environmental samples with
metagenome-assembled genomes and showed that individual microbes within a community have
differing growth requirements. This work guides identification of growth constraints for laboratory
cultivation of diverse microbes.
Code and Data availability
Code is available as the python package GenomeSPOT (Genome-based Salinity, pH, Oxygen
Tolerance, and Temperature for Bacteria and Archaea) at github.com/cultivarium/GenomeSPOT.
Data used in this research is publicly available at BacDive, NCBI GenBank, JGI Integrated Microbial
Genomes & Microbiomes, and the Genome Taxonomy Database (GTDB).
Barnum et al. Page 1 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Introduction
In order to grow a microorganism in the laboratory, its metabolic (nutrition and energy) and
physicochemical (temperature, pH, salinity and oxygen) requirements must be met. Due to the vast
combinatorial search space of possible chemical and physical parameters, robust conditions for
culturing microbes are laborious to determine
1–3
. Computational predictions that could minimize the
amount of experimental labor by constraining this search space would be highly desirable. Unlike
metabolic requirements that can sometimes be predicted from known genes and pathways
4–8
,
physicochemical requirements often lack known genetic determinants and are more difficult to
predict.
Current methods to predict temperature, pH, salinity, and oxygen tolerance may not be broadly
extensible to uncultivated microbes. Most microbes have only few experimentally characterized
close relatives, limiting methods that rely on empirical data for phylogenetic relatives
9
. An alternative
strategy is to correlate operational taxonomic units (OTUs) to physicochemical variables like pH and
oxygen across habitats
10
. It is unclear if gene-based models can be more accurate
1,11–16
. First, the
genes underlying these physicochemical adaptations in diverse organisms remain unknown.
Second, most existing gene-based models for prediction of microbial growth conditions rely on
phylogenetically-skewed databases for their training, leading to inaccurate predictions for new
taxa
17,18
. To build predictive models for uncultivated microbes, careful consideration of phylogenetic
bias is required. This includes training on phylogenetically balanced datasets, evaluation with
phylogenetically novel examples (“out-of-clade prediction”), and using features that are less prone
to spurious phylogenetic correlation
18
.
We hypothesized that models trained on features of DNA and protein sequences from microbes
with known growth conditions could provide more robust and less phylogenetically-biased
predictions for growth of uncultivated microbes. This is because changes in amino acid frequencies
across an entire genome are less likely than losing or adding gene content, and thus more likely to
be predictive across diverse organisms. For example, while some bacterial species can evolve to
grow faster through the loss of many different genes
19
, maximum growth rate across diverse phyla
is consistently correlated with bias in codon usage
20
. For oxygen sensitivity, aerobic organisms tend
to have higher G+C content compared to anaerobic relatives, and amino acid frequencies have
been linked to oxygen use
16,21–23
. For temperature, proteins tend to contain more aromatic and
hydrophobic residues at higher temperatures
24–26
. For salinity, acidic amino acids and lower protein
isoelectric points (pI) tend to be more frequent at higher salinity
27–29
. There have been no reported
correlations of sequence composition with pH, possibly because the vast majority of proteins are
intracellular and the cytoplasm remains close to neutral pH
30
.
Here, we evaluated the ability of models trained on properties of DNA and protein sequences to
predict temperature, pH, salinity, and oxygen preferences of phylogenetically novel microorganisms
Barnum et al. Page 2 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

(Fig. 1A). Models were trained on phylogenetically balanced data derived from 15,596 microbes
and their performance was assessed using phylogenetically constrained out-of-clade testing. We
then used the model to predict the cultivation requirements of 85,205 sequenced species of
Bacteria and Archaea and metagenome-assembled genomes from 3,349 habitats with different
physicochemical conditions.
Results
Overall model design
We chose to predict optimal growth conditions for four physicochemical factors: pH, temperature,
salinity and oxygen tolerance (Fig. 1A). Importantly, we sought to build a predictive model without
relying on gene functional annotations, where the only user input is a genome sequence.
First, we identified a set of DNA and protein sequence features that have been suggested to
correlate with growth conditions (Table 1). For DNA sequences, we included G+C content (DNA
1-mers), frequency of purine-pyrimidine transitions, and estimated coding density. For protein
sequences, we considered protein length, amino acid frequencies (amino acid 1-mers), predicted
isoelectric point (pI), hydrophobicity, number of thermostable residues, hydration state (nH2O),
average carbon oxidation state (Zc), and proportion of arginine to lysine residues (R/R+K). In
addition, we considered the frequency of proteins within specific pI ranges (“isoelectric point
frequencies”). We decided to restrict features to 1-mers because using longer k-mers of DNA or
amino acid sequences can introduce spurious phylogenetic correlations due to their intrinsic
phylogenetic signal
31
and their high-dimensionality relative to the amount of training data available
18
.
To detect any potential influence of extracellular salinity and pH on protein composition, we chose
to calculate protein features not only for all cellular proteins but also for subsets of intracellular-,
extracellular-, and membrane-localized proteins. Extracellular proteins were identified with a hidden
Markov model (see Methods) and the numerical difference between extra- and intra- cellular
proteins (“Δ Extra vs. Intra”) were explored for prediction.
Barnum et al. Page 3 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Figure 1. Overview and assessment of approach. (A) Schematic depiction of our approach. Only a fraction of
organisms have reported growth conditions. Circles represent proportion between the number of all genomes in BacDive
(gray), organisms in BacDive with empirical description of oxygen tolerance (dark blue), and organisms in BacDive with
Barnum et al. Page 4 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

empirical description of temperature, salinity or pH (light blue). We hypothesized properties of DNA and protein
sequences that can be used to predict temperature, pH, salinity, and oxygen. Our approach was to obtain and curate a
dataset of growth conditions, explore their correlations to genome properties, and evaluate the ability of models to
predict growth conditions for novel groups of organisms. Finally, we demonstrated potential applications of such models
by expanding our prediction from the BacDive dataset (represented by gray circle) to all sequenced organisms
(represented by yellow circle), totalling 85,205 prokaryotic genomes. Circle size is proportional to database size. (B)
Distribution of curated data available for cultivated organisms as found in the BacDive dataset. (C) Correlation between
physicochemical growth conditions and genomic features, as calculated for all genomes in the curated dataset. Growth
conditions refer to optimum conditions unless otherwise noted. Each row represents one genome feature (refer to Table
1 for feature descriptions). Color scale represents the degree of correlation: red indicates positive Spearman correlation
coe
fficient, blue indicates negative). DNA sequence features were calculated for the whole genome. Protein sequence
features were calculated based on predicted cellular localization of the resulting proteins. ‘Intracellular’ / ‘Extracellular’ /
‘Membrane’ represent values computed only on proteins with those localizations. ‘No localization’ represents calculation
for all proteins regardless of predicted localization. ‘Δ Extra. vs. Intra.’ represents the di
fference between values computed
on those sets. Black boxes indicate selected correlations shown in panel D. (D) Example of signi
ficant correlations
observed between genetic features and growth conditions in panel C.
Next, we obtained empirical data describing abiotic conditions for 15,596 sequenced microbes
from the BacDive database
1
. To utilize phenotypic data in our model we performed the following
adjustments (see Methods): for temperature, pH, and salinity, we ignored qualitative parameters
(e.g. “thermophile”) and filtered for precise measurements by only using continuous numerical
parameters that have multiple measured values (e.g. a microbe with a 30°C optimum is ignored if
no other temperatures were tested). We also chose to represent oxygen tolerance as probability,
assigning ‘1’ for organisms described as ‘obligate aerobe’, ‘aerobe’, ‘facultative anaerobe’, or
‘facultative aerobe’ and ‘0’ to organisms described as ‘obligate anaerobes’ or ‘anaerobes’ without
other labels. After curation, the number of bacteria and archaea available were 7293 for oxygen
tolerance, 2418 for temperature, 801 for salinity, and 1020 for pH.
The distribution of BacDive data after curation is summarized for each growth condition in Figure
1B. Curation of quantitative values removed some bias, but distributions remain imbalanced: 77%
of organisms were labeled oxygen tolerant, 87% of organisms are mesophiles (15-45°C), 74%
have an optimum salinity between 0-5% w/v NaCl, and 65% have an optimum pH between 6 and
8. Table 2 summarizes the number of genomes and the range of values that were used for each
condition.
Correlation between sequence features and optimum physicochemical conditions
To assess the prospect for successful modeling, we calculated the above sequence features for
each BacDive genome (Supp. Data 1), and measured their correlation with oxygen tolerance,
temperature, salinity, and pH (Fig. 1C). No strong correlation was observed using DNA sequence
features. We observed several strong correlations using protein sequence features. In several
instances we observed stronger correlation when protein localization was considered, likely
explained by differences between intracellular and extracellular salinity and pH. For example,
optimum temperature correlated with the overall frequency of glutamic acid (Spearman correlation,
Barnum et al. Page 5 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

ρ=0.39), optimum salinity correlated with the frequency of aspartic acid among extracellular
proteins (ρ=0.69), optimum pH correlated with the difference between glutamic acid frequency for
extracellular and intracellular proteins (ρ=0.56), and oxygen tolerance was correlated with low
overall cysteine frequency (ρ=-0.49) (Fig. 1D). Remarkably, 69% of anaerobes but only 7% of
oxygen tolerant microbes have more than 1.1% cysteine on average. Strong correlations were also
observed between oxygen tolerance and the frequency of other amino acids. For example, across
most protein localizations, the frequency of tryptophan, histidine, and alanine are positively
correlated with oxygen tolerance, while frequency of cysteine, glutamic acid, and tyrosine are
negatively correlated.
Optimal physicochemical conditions can be predicted solely from sequence
We next developed models to predict growth conditions for novel taxa from sequence features. For
each growth condition, we tried several estimators (untrained models) and nine different sets of
features, grouped by feature type and protein localization. We assessed the performance of each
model (an estimator trained on a set of features) through 5-fold cross-validation with holdouts at
the family level (Supp. Fig. 1, Supp. Data 2, Supp. Data 3). We selected a single model for each
growth condition based primarily on performance but also preferring simpler estimators, fewer
features, and more interpretable features.
The accuracy of the selected models is summarized in Figure 2A and Table 3. Overall, we
assessed oxygen tolerance to have the highest prediction accuracy (F1=0.94), followed by salinity
(R
2
=0.81, RMSE=2.8% w/v NaCl) and temperature (R
2
=0.73, RMSE=6.5 °C). Optimal pH was least
accurately predicted overall (R
2
=0.48, RMSE=1.1 pH). We found that amino acid frequencies alone
were sufficient to provide all four predictions (Fig. 2B). Protein localization slightly increased the
accuracy of predictions for optimum temperature and salinity (Supp. Fig. 1B-C) and was critical for
predicting optimum pH (Fig. 2B). Notably, isoelectric point frequencies could accurately predict
optimum salinity but not other conditions. In all models, minima and maxima were predicted with
roughly the same accuracy as optima when the same estimator and features were used (Supp.
Fig. 2). When challenged with a test dataset composed of families not in the training dataset,
model performance was consistent with training and cross-validation data yet with lower accuracy
(Table 3). This difference is due either to the smaller size and distinct composition of the dataset or
to overfitting during model selection. We expect these models to be predictive on novel families to
the degree of average error (RMSE) between the test and cross-validation data (Fig. 2A, Table 3).
Barnum et al. Page 6 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Figure 2. Accuracy of predictive models. (A) Accuracy of predictions for the selected model (estimator and features).
Refer to Table 3 for the estimator and features selected for each model. Prediction accuracy on validation (black) and
test sets (blue) for optimum temperature, pH, and salinity are shown. Dashed gray lines indicate the baseline prediction,
and solid gray lines indicate perfect predictions for regressions. Holdouts for the cross-validation and test datasets were
performed at the family level, which means that the results re
flect accuracy on new examples of families. (B)
Performances during model selection when the estimator is held constant and the features are varied. Performance was
evaluated in terms of coe
fficient of determination (R
2
) for regression or F1 score (F1) for classification. Negative R
2
values,
which result from random or otherwise less accurate prediction than selecting the mean, are not displayed. The types of
Barnum et al. Page 7 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

features include: “AAs” amino acid frequencies; “pIs” isoelectric point frequencies; “Other” any feature not included in
“AAs” or “pIs”; “All” all features. The localization of features can be: “No Localization” features were computed without
considering localization; “Extra., Intra., Membrane” protein features were calculated on groups of proteins with the same
localization; and “Δ Extra. Vs. Intra.” protein features were calculated as the di
fference between the values for extracellular
and intracellular proteins. Dashed gray line represents the baseline for linear regression (always predicting the mean) or
for logistic regression (always predicting the mode). (C) In
fluence of taxonomic holdout level on model performance.
Black line represents the selected model evaluated using cross-validations performed at di
fferent taxonomic levels. Dark
red line represents an alternative method provided for comparison, where the prediction is the average values of the
closest relatives (e.g. when the holdout level is genus, the closest relatives are from other genera in the family at best).
Light red line represents the same method but a random relative is chosen instead of using the average. Dashed gray line
represents the baseline. Gray highlights indicate areas in which all methods tended to perform better than baseline. (D)
Relationship between accuracy and genome completeness. Black dot is the mean error from 100% completeness and
gray lines are ±1 standard deviation. Error was calculated using 20 genomes evenly distributed across percentiles of
predicted values.
We further investigated the prediction of oxygen tolerance from amino acid frequencies. Different
amino acid frequencies between aerobes and anaerobes have been noted in early comparative
genomics analyses
22
. Such differences could be due to phylogenetic confounders, as several large
phyla are predominantly aerobes or anaerobes and differ in G+C content. Yet the model is able to
discriminate between aerobes and anaerobes with a balanced accuracy of >90% from 35% to
65% G+C content, with 70% balanced accuracy outside of this range (Supp. Fig. 3A). The model
also accurately predicts aerobes and anaerobes across prokaryotic phyla regardless of if they were
composed primarily of aerobes or of anaerobes (mean 90% accuracy across phyla) (Supp. Fig.
3A). When the model was trained on a single phylum with equal proportions of aerobes and
obligate anaerobes, Bacteroidota (n=419 genomes), predictions remained accurate for other phyla
(F1=0.92) (Supp. Fig. 3B). Even two amino acids – cysteine and histidine, tryptophan, glutamate,
or glutamine – can predict oxygen tolerance accurately (F1>0.90) (Supp. Fig. 4). Overall, this
simple logistic regression model based on amino acid frequencies, which applies one set of rules
across all organisms, provides a very accurate prediction (92% balanced accuracy) of oxygen
tolerance generalizable to almost all bacteria and archaea.
We observed model predictions to be less accurate in some ranges of growth conditions. Optimum
temperature was poorly predicted below 15°C (RMSE 14°C in cross-validation) (Fig. 2A). Optimum
pH was systematically overpredicted below pH 5 and underpredicted above pH 9.5 (RMSE 2.0
and 1.8) (Fig. 2A), meaning the true pH optimum in those ranges may be more extreme than
predicted. Optimum salinity was poorly predicted at 10-20% w/v NaCl for organisms that are not
“salt-in” halophiles with extremely acidic proteins (RMSE 4.8% w/v NaCl) (Fig. 2A). At extreme
growth conditions, such as temperatures ranging 60-80°C or salinity above 15%, the shift in amino
acid frequencies results in a less accurate prediction of oxygen tolerance (balanced accuracy of
75% and 67%, respectively) (Supp. Fig. 3A).
The oxygen model contradicted reported classifications which warrant further investigation. Genera
with a mix of aerobes and anaerobes accounted for 26% of instances where the oxygen tolerance
Barnum et al. Page 8 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

model was incorrect. Specific pathways such as anoxygenic phototrophs and anaerobic sulfur
oxidizing bacteria were often incorrectly predicted to be aerobes. Specific phyla such as
Campylobacterota and Chloroflexota were also less accurately predicted. We hypothesize that the
source of these inconsistencies may be due to inaccurate literature reports or exceptions to this
model. Follow-up studies to examine the relationship between amino acid frequencies and
metabolic niches will be required.
Predictive models are robust to phylogenetic bias
Next, we evaluated the influence of phylogenetic bias on all models. A high degree of phylogenetic
bias will result in highly accurate prediction for related species (e.g. in the same genus or family)
and less accurate prediction for distant relatives (e.g. phylum and class). We repeated model
training and evaluation using holdouts at increasing taxonomic ranks. Our results show that
phylogeny had a relatively small effect on the accuracy of our sequence-based models for oxygen
tolerance, temperature, salinity, and pH (Fig. 2C).
As a point of comparison, we show that using the same data, models strongly influenced by
phylogeny fail to predict growth parameters at higher taxonomic ranks and are only accurate for
close relatives (Fig. 2C, Supp. Fig. 5). These models, which strongly depend on the composition
of the training dataset and the nature of phylogenetic correlation, are therefore more useful for
interpolation (predictions for taxa like those in model training) than extrapolation (predictions for
new, different taxa). For example, the phylogeny-based model of oxygen tolerance retained a
moderate overall accuracy up to order and even class levels (Fig. 2C) because of the conservation
of oxygen tolerance across large clades of organisms
2
and the presence of many organisms in
those clades in the dataset. Overall, these results highlight the importance of holding out test data
at higher taxonomic levels to evaluate model performance
18,32
. The accuracy of our models on
higher taxonomic ranks supports their use on phylogenetically novel organisms.
Prediction using incomplete genomes
Models based solely on sequence features require a representative portion of a genome to provide
accurate predictions, and can thus be applied to partial genome sequences. This is particularly
meaningful for metagenome-assembled genomes, which can often be incomplete, and for
genomes with non-standard genetic codes, where protein predictions may be incorrectly truncated
To determine the effect of genome completeness on model performance, we randomly
subsampled the proteins and contiguous fragments of a genome, independently, to 10-100%
completeness. We performed this test on 20 different species spanning each condition’s range and
found that our models, other than pH, retained an acceptable accuracy down to 10% genome
completeness (Fig. 2D, Supp. Fig. 6). Specifically, we found the mean difference between 10%
and 100% genome completeness to be negligible for oxygen tolerance (change in probability of
0.02), 4°C temperature, and 0.7% w/v NaCl salinity. Prediction of pH was substantially less
Barnum et al. Page 9 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

accurate, with an error of 0.4 pH units at 10% genome completeness. Overall our models are
compatible with the 50% completeness threshold used by many metagenome-assembled genome
studies and the Genome Taxonomy Database
33
.
Prediction of physicochemical conditions for all sequenced bacteria and archaea
Of the 85,205 sequenced species of bacteria and archaea available, an estimated 70% are
uncultivated
33
. Uncultivated organisms were not present in our training dataset and may have
unpredictable differences compared with cultivated organisms. We thus evaluated model
performance on uncultivated organisms, specifically new taxonomic groups and less-complete,
metagenome-assembled genomes. We first quantified the similarity of predictions for cultivated
and uncultivated organisms in the same taxonomic family. We reasoned that at the family level,
species might differ in cultivability while typically preferring similar growth conditions. We compared
all 820 families with both cultivated and uncultivated species in the Genome Taxonomy Database,
where the average family consists of 62% uncultivated species (range between 0.6-99.7%). We
found high correlation (R
2
=0.8-0.9) between predicted values for cultured and uncultured microbes
across all models (Fig. 3A), suggesting that our model can robustly predict growth conditions for
uncultivated species. The high correlation for oxygen (R
2
=0.91) and relatively low correlation for pH
(R
2
=0.79) agree with the expected phylogenetic conservation of those growth conditions
2,11
.
We found several examples of taxonomically distinct lineages suggesting our results are in
agreement with previously published observations. The oxygen tolerance model correctly predicted
anaerobic classes of Actinobacteria and the class Vampirovibrionia (formerly Melainabacteria) in the
phylum Cyanobacteriota to be anaerobic, unlike its aerobic relatives (Supp. Fig. 7)
34,35
. Within the
Chloroflexota class Dehalococcoidia, the model correctly identified the order Dehalococcoidales to
be entirely anaerobic and orders Tepidiformales/OLB14 and SAR202/UBA1151 as containing
aerobes, in agreement with previous reports
36–38
. Elevated optimal temperature was correctly
predicted for phyla Hadarchaeota and Calescibacterota (Fig. 3B)
39,40
. Elevated salinity was
correctly predicted for the phylums Nanohaloarchaeota (Fig. 3B)
41
and
Salsurabacteriota/T1Sed10-126 (Supp. Data 4), which additionally has a predicted optimum pH in
line with its reported habitat in soda lakes
42,43
. In addition, a large proportion of species in the phyla
Eremiobacteriota, SZUA-79 (monotypic order Acidulodesulfobacterales), and Micrarchaeota were
predicted to grow at low pH (Fig. 3B), consistent with previous assessments
10,44,45
. These
examples support our hypothesis that sequence feature-based models can learn from cultivated
microbes and be generalized to predict physicochemical requirements for uncultivated groups of
life.
Barnum et al. Page 10 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Figure 3. Predicted physicochemical growth conditions for 85,205 sequenced species of Bacteria and
Archaea. (A) Evaluation of prediction bias between cultivated (x-axis) and uncultivated (y-axis) species. Correlation is
shown between the average value for cultivated vs. uncultivated species in the same family, reported in coe
fficient of
determination. Most values for oxygen tolerance are near 0 and 1. (B) Proportion of species predicted to grow at a given
growth condition. Each plot represents one physicochemical growth condition. Each row on the y-axis represents a
phylum with 10 or more sequenced species, shown in phylogenetic order starting with Archaea. The x-axis represents
percent of all tested species. Each color represents a bin of values for this growth condition (e.g. “pH 5” is an optimum
from pH 5-6 and “Oxygen p>0.5” are predicted aerobes). (C) Comparison between cultivated and uncultivated
organisms predicted to grow at extreme growth conditions. For each physicochemical parameter (y-axis), the fold change
(log2 scale) between cultivated and uncultivated species is shown. Positive values indicate the proportion of uncultivated
species with those conditions is greater than the proportion of cultivated species. Color indicates results for di
fferent
groups of uncultivated species: black - all uncultivated species; light blue - uncultivated species in families with at least
one cultivated representative; dark blue - uncultivated species in families with no cultivated relatives (i.e. uncultivated
above the family level).
Barnum et al. Page 11 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

We used the models to compare overall growth conditions between uncultivated microorganisms
and those cultivated thus far (Fig. 3C, Supp. Fig. 8). Perhaps unsurprisingly, we found that
uncultivated organisms are more likely to be thermophiles, anaerobes, and acidophiles. Specifically,
uncultivated organisms were 3.5-fold more likely to grow at temperatures between 45-60°C,
3.3-fold more likely to grow in anoxic conditions, and 2.6-fold more likely to grow at pH below 5.
Anaerobes accounted for 54% of uncultivated species but only 16% of cultivated species (Supp.
Data 4). Microorganisms in families lacking any cultivated members were 4.1-fold more likely to be
thermophiles relative to uncultivated microorganisms with cultivated relatives (Fig. 3C). For
example, 18% of species in uncultivated families were predicted to grow optimally between
45-60°C, compared to only 4% of species in cultivated families (Supp. Data 4). While the census
of microbial life from metagenomes is incomplete and biased, these data support the notion that
successful cultivation of diverse lineages may require development of accessible laboratory
methods for manipulation at a broad range of growth conditions.
Prediction of physicochemical conditions for metagenomes across habitats
A key consideration for cultivating microorganisms from a given habitat is the design of a laboratory
environment to support their growth. Often, growth conditions are chosen that closely resemble the
microorganism’s habitat. In order to assess the heterogeneity of growth conditions between and
within organisms sharing the same habitat, we predicted optimum growth conditions for a publicly
available dataset of 52,515 metagenome–assembled genomes from various habitats (Supp. Data
5)
46
. We compared all 3,349 metagenomes that contain five or more genomes, of which 37% are
from vertebrate hosts, 21% are from marine environments (includes waters, sediments, and
hydrothermal vents), 12% are from freshwater, and 5% are from soil. We found the average data
across metagenomes generally aligns with expected habitats trends (Fig. 4A). For example,
vertebrate-related metagenomes, predominantly from gut samples, were largely anaerobic and
displayed limited variation in pH (median 7.1) and temperature (median 36°C) (Fig. 4A). As
expected, the median of the average optimum temperatures of thermal springs metagenomes and
thermal marine metagenomes was 69°C and 62°C, respectively. Similarly, metagenomes with a
mean predicted optimum salinity above 6% w/v NaCl, 81% were from non-marine saline and
alkaline or engineered environments. Metagenomes with mean predicted pH of 5.0 or less were
from thermal springs, ferrous or sulfidic biofilms, peatlands, bogs, or “freshwater.” Therefore, our
models can extend laboratory based measurements of microorganisms to estimate environmental
conditions.
Barnum et al. Page 12 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Figure 4. Predicted physicochemical growth conditions for metagenome-assembled genomes from 3,349
metagenomes. (A) Predicted values for each growth condition in each metagenome, with the mean shown as black dot
and ±1 standard deviation shown in gray. Metagenomes (y-axis) were sorted by clustering within each biome (label). The
number of metagenomes in each biome is shown in parentheses. The vertical line is the average values across all
metagenomes in this dataset. (B) Distributions of predicted growth conditions among genomes in each metagenome.
Each color represents a bin of values for this growth condition (e.g. “pH 5” is an optimum from pH 5-6 and “Oxygen
p>0.5” are predicted aerobes). (C) Examples of how combinations of predicted growth conditions can vary for speci
fic
organisms within three metagenomes with 20 metagenome-assembled genomes each. Colors correspond to those used
Barnum et al. Page 13 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

in (B) and text indicates genus and class as assigned in the GEM dataset (some taxonomic levels are unassigned).
Minimum and maximum values for each condition are listed next to each metagenome.
Yet within a metagenome, predictions for individual genomes can vary from the average. For
example, metagenomes with mostly obligate anaerobes, like those from vertebrate guts, often
contained aerobes and metagenomes with mostly aerobes, like those from marine environments,
sometimes contained obligate anaerobes (Fig. 4B). Across all metagenomes, the difference
between the minimum and maximum predicted optima for organisms in a metagenome had a
median value of 16°C, 3.2% NaCl, and 1.6 pH units. Possible explanations for this heterogeneity,
other than prediction error, could include a combination of heterogeneous conditions at the
microscopic scale, environmental fluctuations, transient dispersal, dormant or non-growing cells,
and relic DNA. We anticipate genome-based predictions of growth conditions could help microbial
ecologists understand in more detail ecological processes contributing to heterogeneity of optimal
conditions, such as migration and environmental fluctuation. Because predictions are made on the
genome level, researchers can identify microorganisms in communities with different combinations
of growth conditions (Fig. 4C). Such predictions may suggest the use of several different growth
conditions to isolate more diverse members of the community, or in select cases even help isolate
a specific microorganism by suggesting conditions supporting their distinct growth.
Discussion
This work provides a set of models for predicting physicochemical growth conditions of
microorganisms based solely on genome features. A key aspect of these models is their
robustness to phylogenetic bias, which allows accurate prediction of multiple taxonomic levels
using a microbe’s genome sequence. We also demonstrate robust prediction using partial genome
sequences that meet the standards of metagenomic analysis (>50% complete). This data allows an
unprecedented examination of predicted physicochemical conditions across presently sampled
bacteria and archaea (Supp. Data 4). This is of critical importance for ongoing efforts to cultivate
diverse microorganisms
47,48
. We envision that the physicochemical models provided here will
contribute to cultivation of uncultured organisms, whose characterization in turn will improve the
models. The models could also be used to understand points in the tree of life where organisms
transitioned from one growth condition to another, such as anaerobe-aerobe transitions
16
.
The ability to predict if a microbe is an aerobe or anaerobe based solely on the frequencies of two
or more amino acids is an unexpected finding that warrants further study. Differences in the
frequency of oxygen-sensitive amino acids between aerobes and anaerobes were initially attributed
to reactivity with oxygen, but this hypothesis has been disputed
22,23,49
. We suspect the correlation
between oxygen tolerance and amino acids across phyla (Supp. Fig. 9A) is largely attributable to
the cost of biosynthesis. Tryptophan and histidine have higher energetic costs than similar amino
acids
50
(Supp. Fig. 9B), and would be relatively more costly to synthesize in anoxic environments
Barnum et al. Page 14 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

where less energy is available. Cysteine is significantly more costly in oxic environments because of
the extra energy needed for cells to use the oxidized forms of cysteine and sulfur
51
. Prediction of
facultative anaerobes must be identified from genes associated with anaerobic respiration or
fermentation. Facultative anaerobes and aerobes have similar amino acid usage, indicated by
probability of oxygen tolerance (Supp. Fig. 9C) and similar overall gene content
12
. A recent effort
was unable to accurately predict obligate aerobes, facultative anaerobes, and obligate anaerobes
using amino acid frequencies (balanced accuracy 55%)
21
. The similarity between facultative
anaerobes and aerobes is evidence of the strong selective pressure that oxic conditions exert on
amino acid usage. Studies of organisms where predicted oxygen tolerance differs from these
identified amino-acid usage patterns (e.g. obligate anaerobes predicted to be oxygen tolerant)
would be of particular interest to better understand selective pressures on specific metabolisms or
taxa.
This work represents a significant improvement over existing resources for predicting
physicochemical growth conditions (Table 4). Our models predict all conditions at once and
include optimum, minimum, and maximum. By predicting continuous values, they provide more
precision than models predicting categories (e.g. “thermophile”). As sequence-based models, they
compute more quickly than models involving gene annotations. The composition of all training
datasets are biased entirely by cultivated organisms, but some published models have other biases
that may impact performance: the best performing oxygen model did not include facultative
anaerobes in model training and evaluation
16
, and the best performing optimum pH model was
trained and evaluated specifically on soil and freshwater bacteria
11
. A key concern with many
models with apparent high performance is that they used high-dimensional features prone to
overfitting on training data (e.g. hundred to thousands of genes or k-mers) yet, except for one such
model
16
, did not use out-of-clade test data to make sure predictions remained equally accurate for
organisms in unrelated taxa, which is unlikely. We expect that modeling efforts will continue to
improve and that the simple, validated sequence-based models presented here will provide a
useful foundation for that research.
One issue with all available predictions of optima is that they are likely not precise enough to help
improve laboratory work with cultured microorganisms. The exception would be situations where
the physicochemical conditions used for isolation are not reasonably close to optimum conditions
for growth. When using models, researchers should be aware of the average error as well as
ranges with higher error (for example, below 25°C and at extreme pH values with our models) and
that unusual organisms could have much greater error. Predictions of minimum, maximum and
optimum can disagree when predicted independently (e.g. maximum equal or less than minimum),
as they do for our salinity predictions for 5% of the GTDB dataset. A key source of error is the
imprecision of the training data, which is biased towards particular intervals (e.g. 25°C, 30°C, 37°C,
42°C). Collection of higher-granularity phenotypic data will help improve models and is especially
important to measure for novel taxa that are more likely to have unusual features
52
.
Barnum et al. Page 15 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

The use of sequence features might be improved by incorporating structural information. For
example, protein isoelectric point would be most accurately calculated using residues on the
protein surface but is currently calculated using all residues in the protein sequence. Structural
predictions could also enable the calculation of new, more informative features, such as
interactions between residues influencing temperature stability
53
. Such structural information could
be needed for accurate prediction of low optimum temperatures, where adaptations to increase
protein flexibility are varied
54
. We expect improvements in structural datasets to support more
accurate computational models for microbiology.
Future efforts can build on sequenced-based predictions to make more accurate gene-based
models. A limitation of sequenced-based models is that, relative to changes in genes, changes in
amino acid composition are slow and sometimes subtle. For example, gene gain and loss enable
transitions between freshwater and seawater that take millions years to become evident in amino
acid composition
27
. Similarly, an organism that acquires oxygen-sensitive genes and becomes
oxygen intolerant would initially continue to have the amino acid composition of an oxygen tolerant
organism. Small shifts in optimum pH can have significant impacts in the solubility and protonation
of specific chemicals, and therefore on the fitness effect of specific genes
55–57
, yet result in only
subtle change to amino acid composition. Despite these potential caveats, gene-based models do
not currently provide a better alternative because in most cases the genes underlying growth
conditions in diverse organisms are unknown. A promising approach would be using the
sequence-based predictions of growth conditions now available for all sequenced cultivated and
uncultivated organisms to find phylogenetically robust correlations between conditions and genes.
Such genes would be an appropriate set of features for building accurate gene-based models.
Conclusions
Microbial adaptation to physicochemical growth conditions of oxygen concentration, temperature,
pH, and salinity leave signatures in the DNA and protein sequences of genomes that can be used
to predict optimum conditions. In predicting these conditions, an important confounder is the
phylogenetic bias of growth conditions among related organisms and which organisms have
conditions measured to begin with. The models and approach presented here provide a foundation
for future work predicting physicochemical conditions with more precision. Presently, these models
provide exciting new information about the physicochemical conditions preferred by uncultivated
organisms, possibly aiding in the cultivation of new lineages of life.
Barnum et al. Page 16 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Methods
Data sources and preprocessing
Publicly available growth phenotypes were obtained from BacDive accessed by API in June, 2023
1
.
Data on temperature, pH, salinity, and oxygen tolerance were coded as follows: for continuous
variables, reported optimum was used, and minimum and maximum were defined as the least and
greatest values where growth was described as “positive” and, for salinity only, “inconsistent.” If an
optimum was reported in a range, which was common for pH, we recorded optimum as the
average of that range. Oxygen tolerance was assigned as tolerant for microbes described as
‘obligate aerobe’, ‘aerobe’, ‘facultative anaerobe’, or ‘facultative aerobe’ and intolerant for microbes
described as ‘obligate anaerobes’ or ‘anaerobes’.
Genome accessions were used to download genomes from NCBI GenBank
58
. Additional genomes
for analyses were downloaded from the Joint Genome Institute Genomic Catalog of Earth’s
Microbiomes (GEM) project
46
and from the Genome Taxonomy Database (GTDB) release r214,
which also supplied the taxonomy used here to describe organisms and to identify nearest
taxonomic neighbors
33
. When not available, predicted coding domain sequences (CDS) were
identified from genomes based on open reading frames using the standard genetic code using
Prodigal v.2.6.3 with default settings
59
.
Preprocessing used an automated curation to remove lower quality phenotypic data. For example,
a study that measured temperature in 1°C increments from 20-40°C is of higher quality than a
study that only measured 30°C, 37°C, and 40°C. Preprocessing sought to deplete the dataset of
lower quality phenotypic data like the latter. For commonly measured optima, defined as optima
that accounted for more than 1% of values (e.g. strains with optimum pH 7.0 accounted for 34% of
pH values), data were removed if the minimum or maximum equaled the optimum; if the difference
between the minimum and maximum was less than 1.5 pH units, 10°C, or 1.5% NaCl (unless
salinity was <0.5% NaCl); or if fewer than 4 total points were reported. More extreme values – pH
<4 or >9, salinity >15% NaCl, and temperature <19°C or >44°C – were not removed as we
reasoned these are more likely to be accurately measured. Data was discarded if any of the
optimum, minimum, or maximum were not reported. Due to the unique potential for data entry
errors with salinity, we discarded organisms from the dataset if they were haloarchaea with a
reported optima under 3.7% or if they were reported to have an extreme salinity (above 14% NaCl)
and were obviously incorrect. We expect that inconsistencies in definitions and testing of oxygen
requirements, such as not testing anaerobes for facultative aerobic growth, mean that oxygen
requirement data also contains errors, but these were not addressed.
Barnum et al. Page 17 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Measurement of genomes
Properties of proteins and DNA sequences for each genome (Table 1) were measured using
custom code. Measurements were made on individual predicted mature proteins, which are protein
sequences where the N-terminal methionine and any signal peptide are removed. Protein
measurements included amino acid frequencies, predicted isoelectric point
60
, Grand Average of
Hydropathy (GRAVY)
61
, and average carbon oxidation state (Zc) and stoichiometric hydration state
(nH2O)
62
. Measurements were then aggregated across proteins in the genome. Measurements
made on DNA are more limited: G+C content, the frequency of purine-pyrimidine transitions, and
the estimated coding density. Genomes were discarded if their coding density was below 60%. To
obtain genomes of varying completeness, proteins were randomly sampled and each DNA
sequence (e.g. contig) had a random contiguous portion sampled according to the desired level of
completeness.
The signal peptide prediction model used above was developed to identify exported proteins very
quickly and off-license to support fast use of the models without use limitations. A hidden Markov
model was constructed as described in published methods reported to be accurate at
discriminating the presence or absence of signal peptides
63–65
. The model has states for positions
in the start, N-terminal, hydrophobic, and cut-site regions of SPase I signal peptides, and the
mature protein sequence and was fit using a published dataset
64
. The model runs very quickly and
has high specificity and sensitivity. Only about 10% of proteins are exported, however, so a
threshold was chosen to only allow 20% of proteins in a genome to be false positives, which
captures 60% of exported proteins in the genome (Supp. Fig. 10). Proteins are assigned as a
localization as follows: “membrane” is the protein’s GRAVY is greater than 0.5 the proteome’s
average GRAVY, otherwise “intracellular soluble” if the protein lacks a signal peptide and
“extracellular soluble” if the protein has a signal peptide.
Model dataset construction
To reduce the imbalances in the dataset towards particular groups of groups, organisms from
overrepresented groups were removed. First, the frequency of taxa in the dataset was compared to
the expected frequency of taxa at each taxonomic level. The fold enrichment in the dataset is equal
to the ratio between observed and expected frequencies for each taxonomic level multiplied
together. Then, a random portion of taxa is selected, with the probability of being selected inversely
proportional to their fold enrichment. The result is a smaller dataset more similar to the expected
composition. Here we used the composition of representative species in the Genome Taxonomy
Database as the expected composition and the portion of data removed was 50% for oxygen data
and 33% for every other condition, which had more extensive curation. Extreme values were
retained as in data preprocessing.
Barnum et al. Page 18 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

The partitioning of data into training and test datasets was taxonomically aware. Random families
totaling 20% of the curated, balanced dataset were held out to make a test dataset, and the
remaining 80% of the dataset was for model training and cross-validation. To ensure the test
dataset had a wide range of data, partitioning at the family level was done independently for
extremely low values (<3rd percentile), extremely high values (>97th percentile), and the remaining
values (3rd-97th percentiles). This introduces some data leakage in the sense that the training and
test set can include members from the same families, but the members in the test set have
extreme traits, whereas the members in the training set do not. In cross-validation, random families
consisting of approximately 20% of the training dataset were held out in each fold.
Model evaluation and selection
Different combinations of features and estimators (untrained models) were tested through
cross-validation to select the features and estimator to use to predict each growth condition (Supp.
Data 3). Nine sets of features tested included three types of variables – amino acid frequencies, pI
distributions, other more-derived features, or all features – and three different
compartmentalizations – no compartmentalization, by compartment (e.g. intracellular, extracellular,
and membrane proteins individually), and differences between extracellular and intracellular
proteins. For all models, standard scaling was applied to the features. Feature selection of either
the 20 most correlated features (measured with the F statistic or mutual information statistic) or the
minimum number of features were used for most models. Regression estimators tested were
standard, ridge, and lasso (no feature selection) linear regressions, with varying hyperparameters,
and multi-layer perceptron regressor with two layers and max features per layer of half the available
features (no feature selection). Classifier estimators tested were Gaussian naive Bayes, logistic
regression, and support vector machine (no feature selection) with a linear kernel, with varying
hyperparameters. Each fold of the cross-validation involved its own scaling, feature selection, and
estimator fitting.
Regression model performance was assessed using the coefficient of determination (R
2
) and root
mean square error (RMSE). Classification model performance was assessed on true positives, false
negatives, etc., using specificity, sensitivity, the harmonic mean of those values (F1), and true and
false positive rates. For model selection, the above combinations of models and features were
compared using R
2
or F1 scores on the cross-validation data (Supp. Fig. 1). Among the highest
scoring estimators and sets of features, we selected an estimator and features based on simplicity
and perceived closeness to the biological phenomena. For example, the oxygen tolerance models
were equally accurate when trained on all features or on only amino acids, so we chose to use only
amino acids. Feature importances in the selected models were computed with the permutation
importance (Supp. Fig. 11). After model selection, models were retrained as before but on all
training data at once (no cross-validation), then predictions were made on the final test dataset.
Barnum et al. Page 19 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Model use
A software package is available for users to predict growth conditions for genomes
(github.com/cultivarium/GenomeSPOT). The final models were trained on all curated, balanced
data from both the training and test datasets. The software reports predicted optima, max, and min
and oxygen tolerance. The runtime is approximately 5-10 seconds per genome per CPU.
Author Contributions
T.B., A.C.C., H.H.L., and N.O. conceived of and designed the project. T.B., A.C.C., and M.M.
designed model training and evaluation. T.B. produced all code, data, and results. T.B., A.C.C.,
M.M., P.C., H.H.L., and N.O. analyzed results and wrote the manuscript. All authors have reviewed
the manuscript and approved of its release.
Acknowledgements
We thank the entire Cultivarium team for their support in this work. We are especially grateful to
Elise Ledieu-Dherbécourt for advice and support on the project. We recognize Paul Carini for the
insight that the influence of oxygen amino acid composition may be explained by amino acid
biosynthesis costs. We extend special thanks to James Knight for code support and review and
Julia Leung and Stephanie L. Brumwell for their feedback on the manuscript. Cultivarium
acknowledges support from Schmidt Futures as a Convergent Research Focused Research
Organization (FRO).
Competing Interest Statement
The authors declare no competing interests.
Barnum et al. Page 20 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Tables
Table 1. Description of genomic variables used in this study. For proteins, variables were
measured based on the localization of the protein to the intracellular, extracellular, or membrane
compartments of the cell, the difference between extracellular and intracellular values, or based on
no localization at all. References relate sequence features to growth conditions.
Sequence
feature
Variable
name
Variable definition
Reference
DNA
G+C content
Fraction of bases that are guanine and cytosine
23,24
R>Y transitions
Fraction of bases that transition from a purine to pyrimidine
24
Coding density
Fraction of bases contained within coding sequences, approximated as the total
length of proteins multiplied by three divided by genome length
56
Protein
Length
Mean number of amino acids across proteins
Hydropathy
Mean grand average of hydropathy (GRAVY) across proteins
61
nH
2
O
Mean stoichiometric hydration state, an estimate of the water molecules needed to
compose a protein
62
Zc
Mean average carbon oxidation state, an estimate of the degree of oxidation of
carbon in a protein
62
R/R+K
Ratio of arginine to arginine plus lysine
54
Thermostable
residues
Sum of the amino acids I, V, Y, W, R, E, L reported to provide a correlation with
high temperatures
25
pI
Mean isoelectric point across proteins, computationally estimated for each protein
from the pKa of its amino acids
27,28
pI frequency
Fraction of proteins in the genome with a pI between assigned values, summing to
1
27,28
Amino acid
frequency
Frequency of each amino acid across all proteins
22,25,28,54
Barnum et al. Page 21 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint
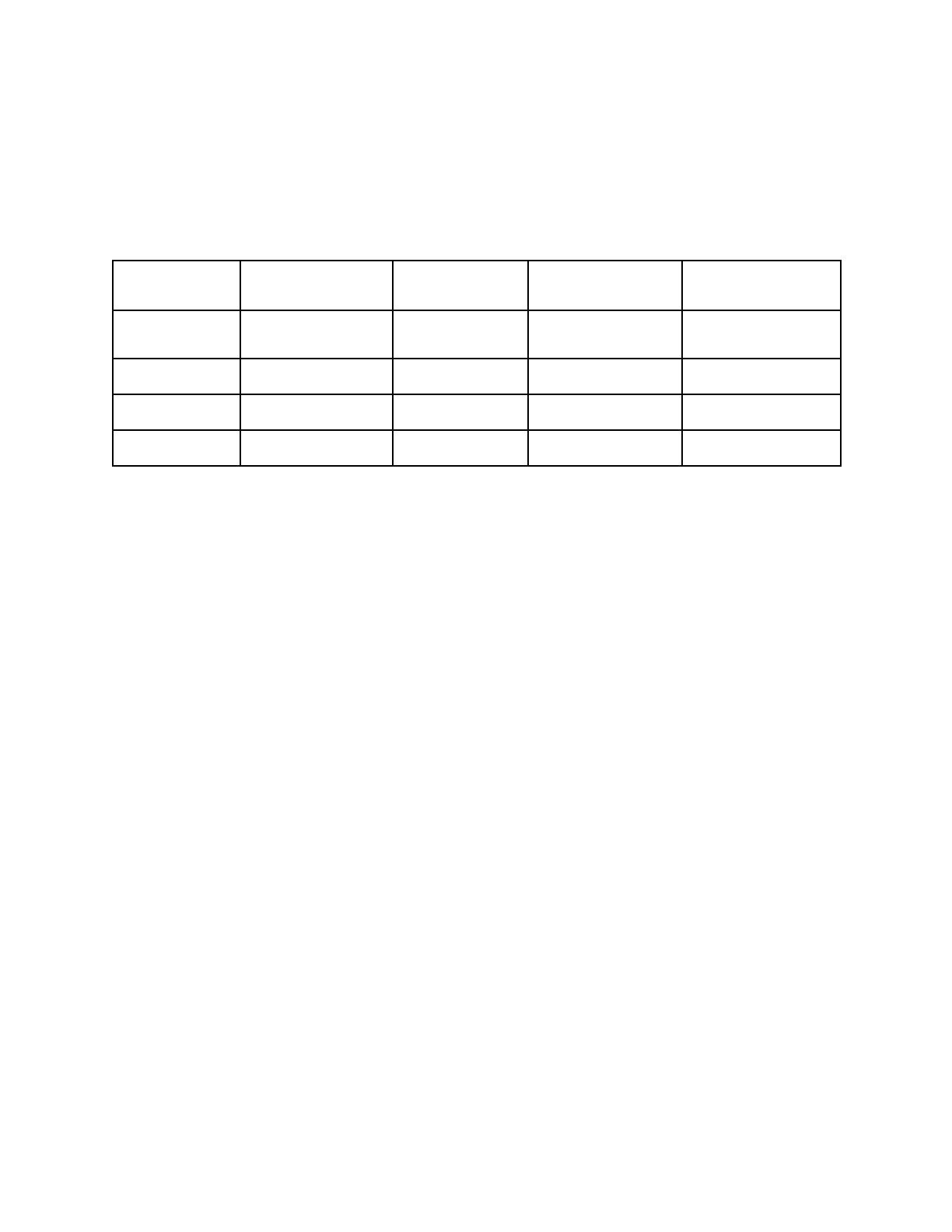
Table 2. Phenotypic data used in this study. Data was obtained from BacDive. The total
number of organisms with optimum values reported is described for (1) all organisms with data and
genomes in BacDive, (2) the subset of those organisms that remained after automated curation,
and (3) the subset of those organisms after a balancing step. The range and mean values for the
organisms used in modeling are provided.
Growth condition
Organisms with data
and genomes
Organisms after
curation
Organisms used in
modeling
Min-Max values in
modeling (Mean)
Oxygen tolerance
7293
7293
3646
0-1 (0.65) probability
tolerant
Temperature
4773
2418
1722
4-105 (33) °C
Salinity
2374
801
593
0.0-27.5 (4.3) % w/v NaCl
pH
3545
1020
756
1.1-12.0 (7.2) pH
Barnum et al. Page 22 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Table 3. Summary of the selected models. The selected features and estimators for each model are indicated. Model performance is
described for four different datasets: training, accuracy of predictions for the training data itself (i.e. fit of the model to its training data);
cross-validation, accuracy of predictions on the examples not used to train the model during each fold of the cross-validation; test,
accuracy of model on held out test data not used for model training or selection; baseline, the accuracy of a baseline prediction, which is
the average of the dataset for regressions and the mode of the dataset for classifiers. Performance can be assessed using the coefficient
of determination (R
2
) and root mean square error (RMSE) for continuous values and the F1 score (F1) for classifications.
Target
Estimator
Training
Cross-Validation
Test
Baseline
Condition
Cardinality
Features
Localization
(Params)
R
2
RMSE
F1
R
2
RMSE
F1
R
2
RMSE
F1
R
2
RMSE
F1
Oxygen Tolerance
Tolerant /
Intolerant
AAs
No Localization
Logistic
Classification
-
-
0.96
-
-
0.94
-
-
0.97
-
-
0.75
Temperature
Optimum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.001)
0.78
5.80
-
0.73
6.53
-
0.72
7.42
-
0.00
12.46
-
Minimum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.001)
0.74
6.65
-
0.68
7.27
-
0.70
8.19
-
0.00
12.94
-
Maximum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.001)
0.79
5.69
-
0.73
6.42
-
0.75
6.74
-
0.00
12.31
-
Salinity
Optimum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.01)
0.87
2.31
-
0.81
2.76
-
0.68
3.26
-
0.00
6.33
-
Minimum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.01)
0.85
1.61
-
0.78
1.93
-
0.64
2.19
-
0.00
4.15
-
Maximum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.01)
0.82
3.82
-
0.72
4.78
-
0.72
4.52
-
0.00
9.08
-
pH
Optimum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.01)
0.61
0.91
-
0.48
1.05
-
0.54
0.89
-
0.00
1.46
-
Minimum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.01)
0.56
0.86
-
0.48
0.94
-
0.49
0.99
-
0.00
1.30
-
Maximum
AAs
Extracellular,
Intracellular, Membrane
Lasso Regression
(alpha=0.01)
0.55
1.14
-
0.35
1.37
-
0.40
1.15
-
0.00
1.70
-
Barnum et al. Page 23 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Table 4. Comparison of published models predicting oxygen, temperature, salinity, and pH. The following parameters are shown
for each study: prediction type; cardinality, the number of things being predicted; features, the variables used for the prediction (k refers
to k-mer size); training data composition, what organisms were used (bias); feature size, the number of features; training data size, the
number of examples learned; balanced taxonomically, whether the publication reports intentionally reducing taxonomic bias to a
significant degree (not simply de-replicating to 1 genome per species); tested, if the accuracy is measured using examples not used in
model training and selection; test out-of-clade, if taxonomic groups were intentionally held out for testing; test accuracy, score to test
performance; accuracy metric, metric used to score performance - accuracy is fraction correct assignments, balanced accuracy is mean
of recall for all predicted categories, F1 is harmonic mean of the precision and recall, R
2
is coefficient of determination; study, publication.
Condition
Prediction
Type
Cardinality
Features
Training data
composition
Feature
size
Training
data size
Balanced
Tested
Tested
out-of-clade
Test
accuracy
Accuracy
metric
Study
Oxygen
Category
Aerobe (Any) or
Obligate Anaerobe
Protein (k=1)
Isolates
20
3646
Yes
Yes
Yes (Family)
0.91
Balanced
accuracy
This work
Oxygen
Category
Aerobe, Facultative
Anaerobe, or
Obligate Anaerobe
Protein (k=3)
Isolates
8000
3350
No
Yes
No
0.7
Balanced
accuracy
Flamholz
21
Oxygen
Category
Aerobe, Facultative
Anaerobe, or
Obligate Anaerobe
DNA (k=5)
Isolates
512
3350
No
Yes
No
0.67
Balanced
accuracy
Flamholz
21
Oxygen
Category
Aerobe or Anaerobe
Genes
Isolates, aerobes
and anaerobes
3507
~3200
No
Yes
Yes (Family)
0.97
Accuracy
Davin
16
Oxygen
Category
Aerobe or Anaerobe
Sequence
alignment
Isolates, aerobes
and anaerobes
99
333
Yes
Yes
No
0.84
Accuracy
Davin
16
Oxygen
Category
Aerobe or Anaerobe
Genes
Isolates
542
5194
No
Yes
No
0.91
Balanced
accuracy
Reimer
1
Oxygen
Category
Aerobe, Facultative
Anaerobe, or
Obligate Anaerobe
Genes
Isolates
325
272
Yes
Yes
No
0.8
Accuracy
Jabłońska
12
Oxygen
Category
Aerobe, Facultative
Anaerobe, or
Obligate Anaerobe
Genes
Isolates
44498
549
No
Yes
No
0.82
Accuracy
Edirisinghe
14
Barnum et al. Page 24 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Condition
Prediction
Type
Cardinality
Features
Training data
composition
Feature
size
Training
data size
Balanced
Tested
Tested
out-of-clade
Test
accuracy
Accuracy
metric
Study
Oxygen
Category
Aerobe, Facultative
Anaerobe, or
Obligate Anaerobe
Genes
Isolates
6554
2231
No
Yes
No
0.86
F1
Weber Zendrera
15
Oxygen
Category
Aerobe or Anaerobe
Genes
Isolates, mostly
pathogens
14831
234
No
Yes
No
~0.97
Accuracy
Weimann
13
pH
Value
Opt., Min., and Max.
Protein (k=1)
Isolates
60
756
Yes
Yes
Yes (Family)
0.54
R
2
This work
pH
Value
Habitat Preference
Genes
Uncultivated soil
and aquatic
genomes
>335
580
No
Yes
No
0.55
R
2
Ramoneda
11
pH
Category
Acidophile or not
Genes
Isolates
542
2710
No
Yes
No
0.67
Balanced
accuracy
Reimer
1
Salinity
Value
Opt., Min., and Max.
Protein (k=1)
Isolates
60
593
Yes
Yes
Yes (Family)
0.68
R
2
This work
Salinity
Category
Halophile or not
Genes
Isolates
542
1162
No
Yes
No
0.98
Balanced
accuracy
Reimer
1
Temperature
Value
Opt., Min., and Max.
Protein (k=1)
Isolates
60
1722
Yes
Yes
Yes (Family)
0.72
R
2
This work
Temperature
Value
Optimum
tRNA (k=1)
Isolates
n.r.
783
No
Yes
Yes (Clusters)
0.59
R
2
Cimen
32
Temperature
Value
Optimum
DNA (k=2-8)
Isolates
~44000
1815
No
Yes
No
0.88
R
2
Wang
66
Temperature
Value
Optimum
Protein (k=2)
Isolates
400
5762
No
Yes
No
0.9
R
2
Li
67
Temperature
Value
Optimum
Protein (k=1)
See Zeldovich
n.r.
n.r.
n.r.
n.r.
Yes
(Metagenomes)
0.87
R
2
Kurokawa
26
Temperature
Value
Optimum
Protein, DNA,
and RNA (k=1)
Isolates
29
2719
No
Yes
No
0.84
R
2
Sauer
24
Temperature
Value
Optimum
Protein (k=1)
Isolates
20
84
No
Yes
No
8-9
RMSE
Zeldovich
25
Temperature
Value
Optimum
Genes
Isolates
6554
782
No
Yes
No
0.77
R
2
Zendrera
15
Temperature
Category
Thermophile or not
Genes
Isolates
542
8406
No
Yes
No
0.79
Balanced
accuracy
Reimer
1
Barnum et al. Page 25 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

Supplementary Information
Supplementary Figure 1. Model selection experiment results for each condition.
Supplementary Figure 2. Performance for minima, optima, and maxima
Supplementary Figure 3. Additional evaluation of the oxygen model.
Supplementary Figure 4. Evaluation of oxygen models using only two features.
Supplementary Figure 5. Holdout experiment scatter plots
Supplementary Figure 6. Accuracy vs. genome completeness for individual genomes.
Supplementary Figure 7. Examples of oxygen classifications for uncultivated lineages.
Supplementary Figure 8. Trait distribution by cultivation status.
Supplementary Figure 9. Understanding the oxygen model.
Supplementary Figure 10. Construction of a hidden Markov model to predict signal peptides.
Supplementary Figure 11. Feature importances in selected models for top 20 features.
Supplementary Data 1. Predictions and reported phenotypes for all BacDive genomes. Columns
titled “reported” refer to phenotypes reported on BacDive. Columns titled “partition” indicate if a
genome was used in the training or test set. Units correspond to those used in the manuscript:
oxygen - the probability of being oxygen tolerant; temperature - degrees Celsius; pH - units; salinity
- % w/v NaCl. GTDB taxonomy is reported. (supplementary_data_1.tsv)
Supplementary Data 2. The number of genomes used in the test, train, or cross-validations (cv)
holdout sets for each physicochemical condition. Data is provided at the family level, which was
used for model selection. (supplementary_data_2.tsv)
Supplementary Data 3. Performance of all features and estimators tested during model selection.
(supplementary_data_3.tsv).
Supplementary Data 4. Predictions for all GTDB representative species and GTDB-provided
taxonomy and NCBI genome category (isolate if “none”). (supplementary_data_4.tsv)
Supplementary Data 5. Predictions for all GEM MAGs and GEM-provided taxonomy and
metagenome metadata. (supplementary_data_5.tsv)
Barnum et al. Page 26 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

References
1. Reimer, L. C. et al. BacDive in 2022: the knowledge base for standardized bacterial and
archaeal data. Nucleic Acids Res. 50, D741–D746 (2022).
2. Barberán, Albert, Caceres Velazquez, Hildamarie, Jones, Stuart & Fierer, Noah. Hiding in Plain
Sight: Mining Bacterial Species Records for Phenotypic Trait Information. mSphere 2,
10.1128/msphere.00237–17 (2017).
3. Lloyd Karen G., Steen Andrew D., Ladau Joshua, Yin Junqi & Crosby Lonnie. Phylogenetically
Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems 3,
10.1128/msystems.00055–18 (2018).
4. Shelton, A. N. et al. Uneven distribution of cobamide biosynthesis and dependence in bacteria
predicted by comparative genomics. ISME J. 13, 789–804 (2019).
5. Dailey, Harry A. et al. Prokaryotic Heme Biosynthesis: Multiple Pathways to a Common
Essential Product. Microbiol. Mol. Biol. Rev. 81, 10.1128/mmbr.00048–16 (2017).
6. Zhou, Z. et al. METABOLIC: high-throughput profiling of microbial genomes for functional
traits, metabolism, biogeochemistry, and community-scale functional networks. Microbiome
10, 33 (2022).
7. Price, M. N., Deutschbauer, A. M. & Arkin, A. P. Filling gaps in bacterial catabolic pathways
with computation and high-throughput genetics. PLoS Genet. 18, e1010156 (2022).
8. Reji, L., Darnajoux, R. & Zhang, X. A genomic view of environmental and life history controls on
microbial nitrogen acquisition strategies. Environ. Microbiol. Rep. (2023)
doi:10.1111/1758-2229.13220.
9. Oberhardt, M. A. et al. Harnessing the landscape of microbial culture media to predict new
organism–media pairings. Nat. Commun. 6, 1–14 (2015).
10. Ji, M. et al. Candidatus Eremiobacterota, a metabolically and phylogenetically diverse
terrestrial phylum with acid-tolerant adaptations. ISME J. 15, 2692–2707 (2021).
11. Ramoneda, J. et al. Building a genome-based understanding of bacterial pH preferences. Sci
Adv 9, eadf8998 (2023).
12. Jabłońska, J. & Tawfik, D. S. The number and type of oxygen-utilizing enzymes indicates
aerobic vs. anaerobic phenotype. Free Radic. Biol. Med. 140, 84–92 (2019).
13. Weimann, A. et al. From Genomes to Phenotypes: Traitar, the Microbial Trait Analyzer.
mSystems 1, (2016).
14. Edirisinghe, J. N. et al. Machine Learning-Driven Phenotype Predictions based on Genome
Annotations. bioRxiv 2023.08.11.552879 (2023) doi:10.1101/2023.08.11.552879.
15. Weber Zendrera, A., Sokolovska, N. & Soula, H. A. Functional prediction of environmental
variables using metabolic networks. Sci. Rep. 11, 12192 (2021).
16. Davín, A. A. et al. An evolutionary timescale for Bacteria calibrated using the Great Oxidation
Event. bioRxiv 2023.08.08.552427 (2023) doi:10.1101/2023.08.08.552427.
17. Bradley, P. H., Nayfach, S. & Pollard, K. S. Phylogeny-corrected identification of microbial gene
families relevant to human gut colonization. PLoS Comput. Biol. 14, e1006242 (2018).
18. Li, Z., Selim, A. & Kuehn, S. Statistical prediction of microbial metabolic traits from genomes.
PLoS Comput. Biol. 19, e1011705 (2023).
19. Price, M. N. et al. Indirect and suboptimal control of gene expression is widespread in
bacteria. Mol. Syst. Biol. 9, 660 (2013).
20. Weissman, J. L., Hou, S. & Fuhrman, J. A. Estimating maximal microbial growth rates from
cultures, metagenomes, and single cells via codon usage patterns. Proceedings of the
Barnum et al. Page 27 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

National Academy of Sciences 118, e2016810118 (2021).
21. Flamholz, A. I. et al. Annotation-free prediction of microbial dioxygen utilization. bioRxiv
2024.01.16.575888 (2024) doi:10.1101/2024.01.16.575888.
22. Naya, H., Romero, H., Zavala, A., Alvarez, B. & Musto, H. Aerobiosis increases the genomic
guanine plus cytosine content (GC%) in prokaryotes. J. Mol. Evol. 55, 260–264 (2002).
23. Aslam, S. et al. Aerobic prokaryotes do not have higher GC contents than anaerobic
prokaryotes, but obligate aerobic prokaryotes have. BMC Evol. Biol. 19, 35 (2019).
24. Sauer, D. B. & Wang, D.-N. Predicting the optimal growth temperatures of prokaryotes using
only genome derived features. Bioinformatics 35, 3224–3231 (2019).
25. Zeldovich, K. B., Berezovsky, I. N. & Shakhnovich, E. I. Protein and DNA sequence
determinants of thermophilic adaptation. PLoS Comput. Biol. 3, e5 (2007).
26. Kurokawa, M. et al. Metagenomic Thermometer. DNA Res. 30, (2023).
27. Jurdzinski, K. T. et al. Large-scale phylogenomics of aquatic bacteria reveal molecular
mechanisms for adaptation to salinity. Sci Adv 9, eadg2059 (2023).
28. Kiraga, J. et al. The relationships between the isoelectric point and: length of proteins,
taxonomy and ecology of organisms. BMC Genomics 8, 163 (2007).
29. Gunde-Cimerman, N., Plemenitaš, A. & Oren, A. Strategies of adaptation of microorganisms of
the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 42, 353–375
(2018).
30. White, D., Drummond, J. T. & Fuqua, C. The Physiology and Biochemistry of Prokaryotes.
(Oxford University Press New York, 2014).
31. Bussi, Y., Kapon, R. & Reich, Z. Large-scale k-mer-based analysis of the informational
properties of genomes, comparative genomics and taxonomy. PLoS One 16, e0258693
(2021).
32. Cimen, E., Jensen, S. E. & Buckler, E. S. Building a tRNA thermometer to estimate microbial
adaptation to temperature. Nucleic Acids Res. 48, 12004–12015 (2020).
33. Parks, D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a
phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic
Acids Res. 50, D785–D794 (2022).
34. Soo, R. M., Hemp, J., Parks, D. H., Fischer, W. W. & Hugenholtz, P. On the origins of oxygenic
photosynthesis and aerobic respiration in Cyanobacteria. Science 355, 1436–1440 (2017).
35. Jiao, J.-Y. et al. Insight into the function and evolution of the Wood–Ljungdahl pathway in
Actinobacteria. ISME J. 15, 3005–3018 (2021).
36. Lim, Y., Seo, J.-H., Giovannoni, S. J., Kang, I. & Cho, J.-C. Cultivation of marine bacteria of
the SAR202 clade. Nat. Commun. 14, 5098 (2023).
37. Löffler, F. E. et al. Dehalococcoides mccartyi gen. nov., sp. nov., obligately
organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation,
belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord.
nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol.
Microbiol. 63, 625–635 (2013).
38. Kochetkova, T. V. et al. Tepidiforma bonchosmolovskayae gen. nov., sp. nov., a moderately
thermophilic Chloroflexi bacterium from a Chukotka hot spring (Arctic, Russia), representing a
novel class, Tepidiformia, which includes the previously uncultivated lineage OLB14. Int. J.
Syst. Evol. Microbiol. 70, 1192–1202 (2020).
39. Becraft, E. D. et al. Single-Cell-Genomics-Facilitated Read Binning of Candidate Phylum EM19
Genomes from Geothermal Spring Metagenomes. Appl. Environ. Microbiol. 82, 992–1003
Barnum et al. Page 28 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

(2016).
40. Baker, B. J. et al. Genomic inference of the metabolism of cosmopolitan subsurface Archaea,
Hadesarchaea. Nat Microbiol 1, 16002 (2016).
41. Narasingarao, P. et al. De novo metagenomic assembly reveals abundant novel major lineage
of Archaea in hypersaline microbial communities. ISME J. 6, 81–93 (2012).
42. Vavourakis, C. D. et al. A metagenomics roadmap to the uncultured genome diversity in
hypersaline soda lake sediments. Microbiome 6, 168 (2018).
43. Gutierrez-Preciado, A. et al. Extremely acidic proteomes and metabolic flexibility in bacteria
and highly diversified archaea thriving in geothermal chaotropic brines. bioRxiv
2024.03.10.584303 (2024) doi:10.1101/2024.03.10.584303.
44. Tan, S. et al. Insights into ecological role of a new deltaproteobacterial order Candidatus
Acidulodesulfobacterales by metagenomics and metatranscriptomics. ISME J. 13, 2044–2057
(2019).
45. Baker, B. J. et al. Lineages of acidophilic archaea revealed by community genomic analysis.
Science 314, 1933–1935 (2006).
46. Nayfach, S. et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 39, 499–509
(2020).
47. Carini Paul. A ‘Cultural’ Renaissance: Genomics Breathes New Life into an Old Craft.
mSystems 4, 10.1128/msystems.00092–19 (2019).
48. Lewis, W. H., Tahon, G., Geesink, P., Sousa, D. Z. & Ettema, T. J. G. Innovations to culturing
the uncultured microbial majority. Nat. Rev. Microbiol. 19, 225–240 (2021).
49. Vieira-Silva, S. & Rocha, E. P. C. An assessment of the impacts of molecular oxygen on the
evolution of proteomes. Mol. Biol. Evol. 25, 1931–1942 (2008).
50. Akashi, H. & Gojobori, T. Metabolic efficiency and amino acid composition in the proteomes of
Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 99, 3695–3700 (2002).
51. Korshunov, S., Imlay, K. R. C. & Imlay, J. A. Cystine import is a valuable but risky process
whose hazards Escherichia coli minimizes by inducing a cysteine exporter. Mol. Microbiol.
113, 22–39 (2020).
52. Grzymski, J. J. & Dussaq, A. M. The significance of nitrogen cost minimization in proteomes of
marine microorganisms. ISME J. 6, 71–80 (2012).
53. Pinney, M. M. et al. Parallel molecular mechanisms for enzyme temperature adaptation.
Science 371, (2021).
54. Bowman, J. P. Genomics of Psychrophilic Bacteria and Archaea. in Psychrophiles: From
Biodiversity to Biotechnology (ed. Margesin, R.) 345–387 (Springer International Publishing,
Cham, 2017).
55. Lund, P., Tramonti, A. & De Biase, D. Coping with low pH: molecular strategies in neutralophilic
bacteria. FEMS Microbiol. Rev. 38, 1091–1125 (2014).
56. Baker-Austin, C. & Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol.
15, 165–171 (2007).
57. Dopson, M. & Holmes, D. S. Metal resistance in acidophilic microorganisms and its
significance for biotechnologies. Appl. Microbiol. Biotechnol. 98, 8133–8144 (2014).
58. Benson, D. A. et al. GenBank. Nucleic Acids Res. 41, D36 (2013).
59. Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site
identification. BMC Bioinformatics 11, 119 (2010).
60. Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular
biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Barnum et al. Page 29 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint

61. Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein.
J. Mol. Biol. 157, 105–132 (1982).
62. Dick, J. M., Yu, M. & Tan, J. Uncovering chemical signatures of salinity gradients through
compositional analysis of protein sequences. Biogeosciences 17, 6145–6162 (2020).
63. Nielsen, H. & Krogh, A. Prediction of signal peptides and signal anchors by a hidden Markov
model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 122–130 (1998).
64. Bagos, P. G., Nikolaou, E. P., Liakopoulos, T. D. & Tsirigos, K. D. Combined prediction of Tat
and Sec signal peptides with hidden Markov models. Bioinformatics 26, 2811–2817 (2010).
65. Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep
neural networks. Nat. Biotechnol. 37, 420–423 (2019).
66. Wang, S. et al. CnnPOGTP: a novel CNN-based predictor for identifying the optimal growth
temperatures of prokaryotes using only genomic k-mers distribution. Bioinformatics 38,
3106–3108 (2022).
67. Li, G., Rabe, K. S., Nielsen, J. & Engqvist, M. K. M. Machine Learning Applied to Predicting
Microorganism Growth Temperatures and Enzyme Catalytic Optima. ACS Synth. Biol. 8,
1411–1420 (2019).
Barnum et al. Page 30 of 30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted March 22, 2024. ; https://doi.org/10.1101/2024.03.22.586313doi: bioRxiv preprint
